2R8Z
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli in complex with a phosphate and a calcium ion | | 分子名称: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CALCIUM ION, PHOSPHATE ION | | 著者 | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | 登録日 | 2007-09-11 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
2R8X
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli | | 分子名称: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CHLORIDE ION | | 著者 | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | 登録日 | 2007-09-11 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | 著者 | Wang, K, Wang, X, Pan, L. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
5WXN
 
 | | Structure of the LKB1 and 14-3-3 complex | | 分子名称: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase STK11 | | 著者 | Ding, S, Shi, Z.B. | | 登録日 | 2017-01-08 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | Structure of the complex of phosphorylated liver kinase B1 and 14-3-3 zeta
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6L1P
 
 | | Crystal structure of PHF20L1 in complex with Hit 1 | | 分子名称: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-29 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.231 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1C
 
 | |
6L1I
 
 | |
5X62
 
 | |
6LAN
 
 | | Structure of CCDC50 and LC3B complex | | 分子名称: | Coiled-coil domain-containing protein 50,Microtubule-associated proteins 1A/1B light chain 3B | | 著者 | Liu, L, Li, J, Hou, P. | | 登録日 | 2019-11-12 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection.
Cell Res., 31, 2021
|
|
6L10
 
 | | PHF20L1 Tudor1 - MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1F
 
 | |
6XZ9
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with 5-chloro-3,3-dimethyl-2-[5-[1-(1-methylpyrazole-4-carbonyl)azetidin-3-yl]oxy-3-pyridyl]isoindolin-1-one | | 分子名称: | 5-chloranyl-3,3-dimethyl-2-[5-[1-(1-methylpyrazol-4-yl)carbonylazetidin-3-yl]oxypyridin-3-yl]isoindol-1-one, Cytochrome P450 11B2, mitochondrial, ... | | 著者 | Kuglstatter, A, Joseph, C, Benz, J. | | 登録日 | 2020-02-03 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
8DTM
 
 | | Cryo-EM structure of insulin receptor (IR) bound with S597 component 2 | | 分子名称: | Insulin mimetic peptide S597 component 2, Insulin receptor | | 著者 | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | 登録日 | 2022-07-26 | | 公開日 | 2022-09-07 | | 最終更新日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
8DTL
 
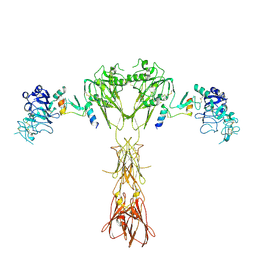 | | Cryo-EM structure of insulin receptor (IR) bound with S597 peptide | | 分子名称: | Insulin mimetic peptide S597, Insulin receptor | | 著者 | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | 登録日 | 2022-07-25 | | 公開日 | 2022-09-07 | | 最終更新日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (5.4 Å) | | 主引用文献 | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
6WBW
 
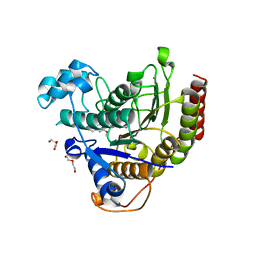 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | 著者 | Klein, D.J, Yu, W. | | 登録日 | 2020-03-27 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6WBZ
 
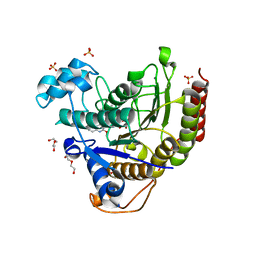 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor containing a spiro-bicyclic group | | 分子名称: | (1S)-N-{(1S)-7,7-dihydroxy-1-[5-(2-methoxyquinolin-3-yl)-1H-imidazol-2-yl]nonyl}-6-ethyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Klein, D.J, Yu, W. | | 登録日 | 2020-03-28 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4N4G
 
 | |
4N4I
 
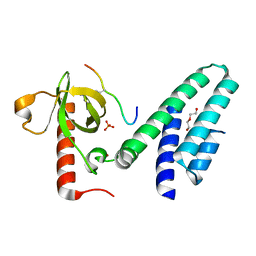 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | 分子名称: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | 著者 | Li, Y, Ren, Y, Li, H. | | 登録日 | 2013-10-08 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
4N4H
 
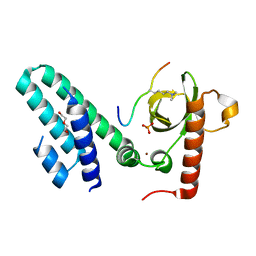 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | 分子名称: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | 著者 | Li, Y, Ren, Y, Li, H. | | 登録日 | 2013-10-08 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
8TH1
 
 | |
7JS8
 
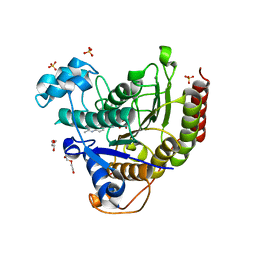 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH AN ETHYL KETONE INHIBITOR CONTAINING A SPIRO-BICYCLIC GROUP (COMPOUND 22) | | 分子名称: | (1S)-N-{(1S)-7,7-dihydroxy-1-[4-(2-methylquinolin-6-yl)-1H-imidazol-2-yl]nonyl}-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Klein, D.J, Yu, W. | | 登録日 | 2020-08-14 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.634 Å) | | 主引用文献 | Discovery of Ethyl Ketone-Based Highly Selective HDACs 1, 2, 3 Inhibitors for HIV Latency Reactivation with Minimum Cellular Potency Serum Shift and Reduced hERG Activity.
J.Med.Chem., 64, 2021
|
|
8TH7
 
 | |
8TH6
 
 | | Crystal Structure of the G3BP1 NTF2-like domain bound to USP10 peptide | | 分子名称: | 1,2-ETHANEDIOL, Ras GTPase-activating protein-binding protein 1, Ubiquitin carboxyl-terminal hydrolase 10 | | 著者 | Hughes, M.P, Taylor, J.P, Yang, Z. | | 登録日 | 2023-07-14 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication and pathogenicity.
Cell Rep, 43, 2024
|
|
8TH5
 
 | |
