1QMA
 
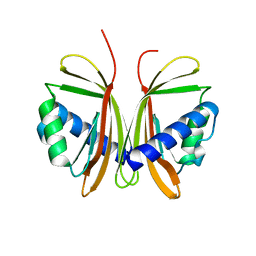 | | Nuclear Transport Factor 2 (NTF2) W7A mutant | | 分子名称: | NUCLEAR TRANSPORT FACTOR 2 | | 著者 | Bayliss, R, Ribbeck, K, Akin, D, Kent, H.M, Feldherr, C.M, Gorlich, D, Stewart, M.J. | | 登録日 | 1999-09-23 | | 公開日 | 2000-02-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Interaction Betweeen Ntf2 and Xfxfg-Containing Nucleoporins is Required to Mediate Nuclear Import of Ran-Gdp
J.Mol.Biol., 293, 1999
|
|
3CW8
 
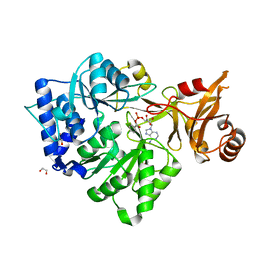 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, bound to 4CBA-Adenylate | | 分子名称: | 1,2-ETHANEDIOL, 4-chlorobenzoyl CoA ligase, 5'-O-[(S)-{[(4-chlorophenyl)carbonyl]oxy}(hydroxy)phosphoryl]adenosine | | 著者 | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | 登録日 | 2008-04-21 | | 公開日 | 2008-09-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
2P4E
 
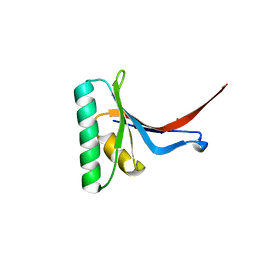 | | Crystal Structure of PCSK9 | | 分子名称: | MERCURY (II) ION, Proprotein convertase subtilisin/kexin type 9 | | 著者 | Cunningham, D, Danley, D.E, Geoghegan, F.K, Griffor, M.C, Hawkins, J.L, Qiu, X. | | 登録日 | 2007-03-12 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1QW9
 
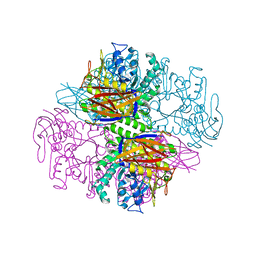 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with 4-nitrophenyl-Ara | | 分子名称: | 4-nitrophenyl alpha-L-arabinofuranoside, Alpha-L-arabinofuranosidase | | 著者 | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | 登録日 | 2003-09-01 | | 公開日 | 2003-10-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1QW8
 
 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with Ara-alpha(1,3)-Xyl | | 分子名称: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose | | 著者 | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | 登録日 | 2003-09-01 | | 公開日 | 2003-10-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
2L31
 
 | | Human PARP-1 zinc finger 2 | | 分子名称: | Poly [ADP-ribose] polymerase 1, ZINC ION | | 著者 | Neuhaus, D, Eustermann, S, Yang, J, Videler, H. | | 登録日 | 2010-08-30 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger.
J.Mol.Biol., 407, 2011
|
|
2L30
 
 | | Human PARP-1 zinc finger 1 | | 分子名称: | Poly [ADP-ribose] polymerase 1, ZINC ION | | 著者 | Neuhaus, D, Eustermann, S, Yang, J, Videler, H. | | 登録日 | 2010-08-30 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger.
J.Mol.Biol., 407, 2011
|
|
7PQY
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FI-3A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FI-3A Fab heavy chain, FI-3A Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-09-20 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
3MHI
 
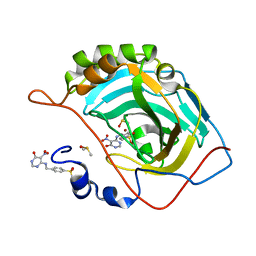 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[(5-nitro-6-oxo-1,6-dihydro-4-pyrimidinyl)amino]methyl}benzenesulfonamide | | 分子名称: | 4-{[(5-nitro-6-oxo-1,6-dihydropyrimidin-4-yl)amino]methyl}benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | 著者 | Grazulis, S, Manakova, E, Golovenko, D. | | 登録日 | 2010-04-08 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
6RC9
 
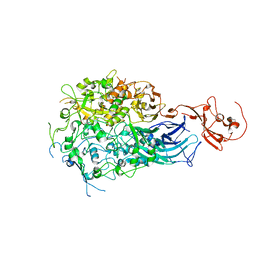 | |
6R8B
 
 | | Escherichia coli AGPase in complex with FBP. | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | 著者 | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | 登録日 | 2019-04-01 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM
Biorxiv, 2020
|
|
6R8U
 
 | | Escherichia coli AGPase in complex with AMP. | | 分子名称: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase | | 著者 | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | 登録日 | 2019-04-02 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM
Biorxiv, 2020
|
|
6O4J
 
 | | Amyloid Beta KLVFFAENVGS 16-26 D23N Iowa mutation | | 分子名称: | Amyloid-beta precursor protein | | 著者 | Griner, S.L, Sawaya, M.R, Rodriguez, J.A, Cascio, D, Gonen, T. | | 登録日 | 2019-02-28 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.402 Å) | | 主引用文献 | Structure based inhibitors of Amyloid Beta core suggest a common interface with Tau.
Elife, 8, 2019
|
|
6RW8
 
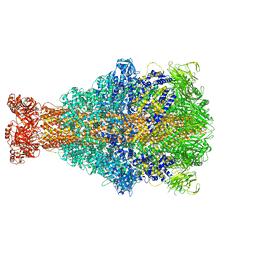 | | Cryo-EM structure of Xenorhabdus nematophila XptA1 | | 分子名称: | A component of insecticidal toxin complex (Tc) | | 著者 | Roderer, D, Leidreiter, F, Gatsogiannis, C, Meusch, D, Benz, R, Raunser, S. | | 登録日 | 2019-06-04 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Common architecture of Tc toxins from human and insect pathogenic bacteria.
Sci Adv, 5, 2019
|
|
1ATT
 
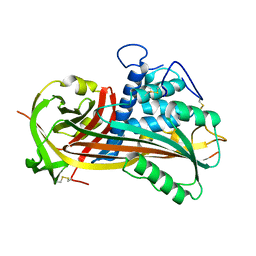 | |
8PEE
 
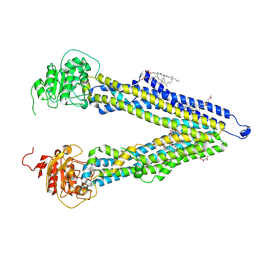 | | ABCB1 L335C mutant (mABCB1) in the inward facing state bound to AAC | | 分子名称: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1, ... | | 著者 | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | 登録日 | 2023-06-13 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
6RWB
 
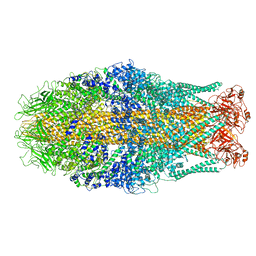 | | Cryo-EM structure of Yersinia pseudotuberculosis TcaA-TcaB | | 分子名称: | Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit,Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit,Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit,Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit | | 著者 | Roderer, D, Leidreiter, F, Gatsogiannis, C, Meusch, D, Benz, R, Raunser, S. | | 登録日 | 2019-06-04 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Common architecture of Tc toxins from human and insect pathogenic bacteria.
Sci Adv, 5, 2019
|
|
8ORQ
 
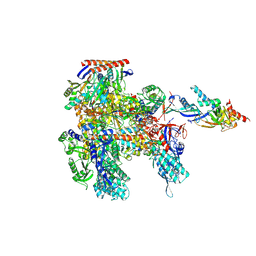 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase open clamp conformation | | 分子名称: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | 著者 | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | 登録日 | 2023-04-17 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
3MYQ
 
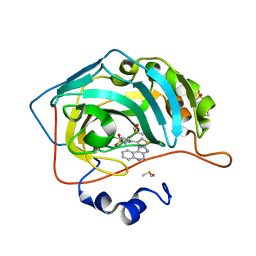 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-[(1H-imidazo[4,5-c]quinolin-2-ylsulfanyl)acetyl]benzenesulfonamide | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-5-[(1H-imidazo[4,5-c]quinolin-2-ylsulfanyl)acetyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | 著者 | Grazulis, S, Manakova, E, Golovenko, D. | | 登録日 | 2010-05-11 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Indapamide-like benzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, and XIII.
Bioorg.Med.Chem., 18, 2010
|
|
7N9C
 
 | |
7N9E
 
 | |
8OUO
 
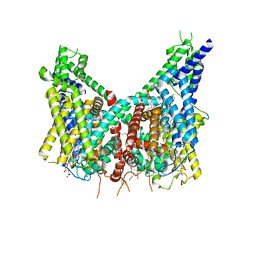 | | Human TPC2 in Complex with Antagonist (S)-SG-094 | | 分子名称: | (1S)-6-methoxy-2-methyl-7-phenoxy-1-[(4-phenoxyphenyl)methyl]-3,4-dihydro-1H-isoquinoline, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | 著者 | Chi, G, Pike, A.C.W, Maclean, E.M, Li, H, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, McKinley, G, Fernandez-Cid, A, Duerr, K. | | 登録日 | 2023-04-24 | | 公開日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for inhibition of the lysosomal two-pore channel TPC2 by a small molecule antagonist.
Structure, 2024
|
|
7N9B
 
 | |
6S19
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.Ch. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1D
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from 20,000 diffraction patterns | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
