8GDB
 
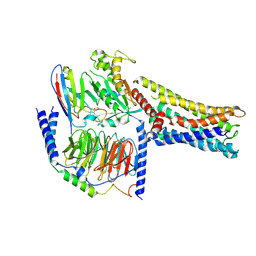 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | 分子名称: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Shenming, H, Mengyao, X, Lei, L, Jianqiang, M, Jinpeng, S. | | 登録日 | 2023-03-03 | | 公開日 | 2024-01-10 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6PUN
 
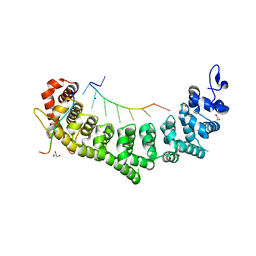 | | Crystal structure of a ternary complex of FBF-2 with LST-1 (site B) and compact FBE RNA | | 分子名称: | 1,2-ETHANEDIOL, Fem-3 mRNA-binding factor 2, GLYCEROL, ... | | 著者 | Qiu, C, Campbell, Z.T, Hall, T.M.T. | | 登録日 | 2019-07-18 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | A crystal structure of a collaborative RNA regulatory complex reveals mechanisms to refine target specificity.
Elife, 8, 2019
|
|
6U0S
 
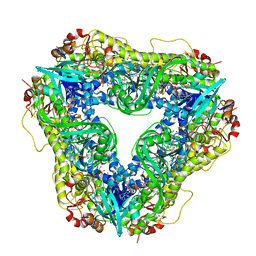 | | Crystal structure of the flavin-dependent monooxygenase PieE in complex with FAD and substrate | | 分子名称: | 2,4-dichlorophenol 6-monooxygenase, 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraen-1-yl]-6-methoxy-3-methylpyridin-4-ol, CHLORIDE ION, ... | | 著者 | Shi, R, Manenda, M. | | 登録日 | 2019-08-14 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
6U0P
 
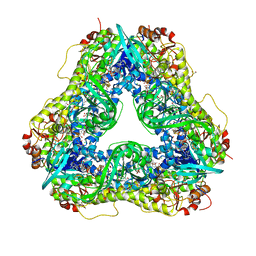 | | Crystal structure of PieE, the flavin-dependent monooxygenase involved in the biosynthesis of piericidin A1 | | 分子名称: | 2,4-dichlorophenol 6-monooxygenase, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Shi, R, Manenda, M, Picard, M.-E. | | 登録日 | 2019-08-14 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
7XPQ
 
 | |
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | 分子名称: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | 著者 | Chen, Y.H, Wang, X.C, Zhang, C.R. | | 登録日 | 2022-05-05 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
7XPP
 
 | |
8JNK
 
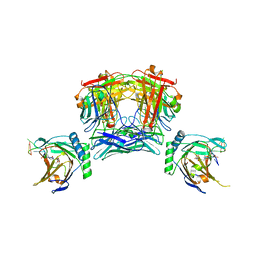 | |
8JNR
 
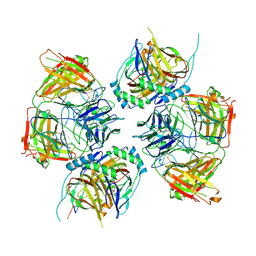 | |
4UX4
 
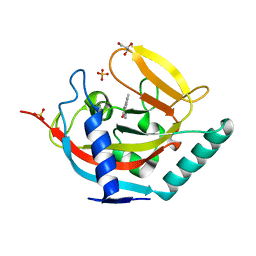 | | Crystal structure of human tankyrase 2 in complex with 1-methyl-7-(4- methylphenyl)-5-oxo-5,6-dihydro-1,6-naphthyridin-1-ium | | 分子名称: | (1S)-1-methyl-7-(4-methylphenyl)-5-oxo-1,5-dihydro-1,6-naphthyridin-1-ium, GLYCEROL, SULFATE ION, ... | | 著者 | Haikarainen, T, Lehtio, L. | | 登録日 | 2014-08-19 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Based Design, Synthesis and Evaluation in Vitro of Arylnaphthyridinones, Arylpyridopyrimidinones and Their Tetrahydro Derivatives as Inhibitors of the Tankyrases.
Bioorg.Med.Chem., 23, 2015
|
|
8TAO
 
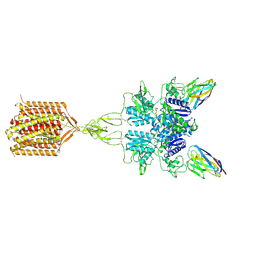 | | Quis and CDPPB bound active mGlu5 | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5, ... | | 著者 | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | 登録日 | 2023-06-27 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8T6J
 
 | |
8T8M
 
 | | Quis-bound intermediate mGlu5 | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nb43 | | 著者 | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | 登録日 | 2023-06-22 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8T7H
 
 | | Quis-bound intermediate mGlu5 | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nanobody 43 | | 著者 | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | 登録日 | 2023-06-20 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
6ASG
 
 | | Crystal structure of Thermus thermophilus RNA polymerase core enzyme | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Liu, Y, Lin, W, Ying, R, Ebright, R.H. | | 登録日 | 2017-08-24 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
5WPW
 
 | |
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | 著者 | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | 登録日 | 2020-07-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.108 Å) | | 主引用文献 | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
5Z4Y
 
 | |
6XEO
 
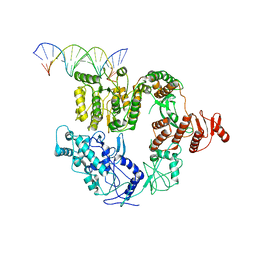 | | Structure of Mfd bound to dsDNA | | 分子名称: | DNA (5'-D(P*AP*GP*GP*AP*TP*AP*CP*TP*TP*AP*CP*AP*GP*CP*CP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*GP*GP*CP*TP*GP*TP*AP*AP*GP*TP*AP*TP*CP*CP*T)-3'), Transcription-repair-coupling factor | | 著者 | Brugger, C, Deaconescu, A. | | 登録日 | 2020-06-12 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | Molecular determinants for dsDNA translocation by the transcription-repair coupling and evolvability factor Mfd.
Nat Commun, 11, 2020
|
|
5Z50
 
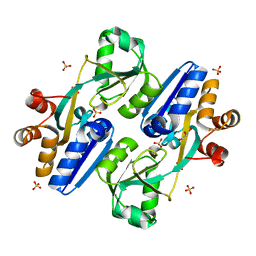 | | Crystal structure of PaCysB regulatory domain | | 分子名称: | Cys regulon transcriptional activator, GLYCEROL, SULFATE ION | | 著者 | Yang, C, Liang, H, Gan, J. | | 登録日 | 2018-01-15 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.206 Å) | | 主引用文献 | Molecular insights into the master regulator CysB-mediated bacterial virulence in Pseudomonas aeruginosa.
Mol.Microbiol., 111, 2019
|
|
5Z4Z
 
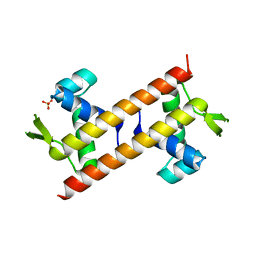 | |
4KXF
 
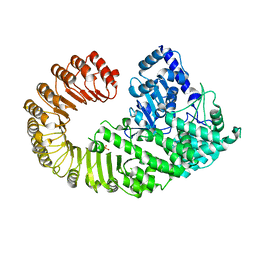 | |
5DDU
 
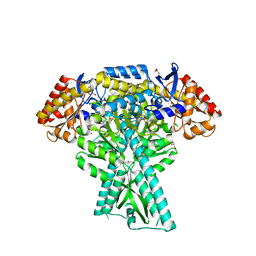 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PMP | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CrmG, GLYCEROL, ... | | 著者 | Xu, J, Feng, Z, Liu, J. | | 登録日 | 2015-08-25 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5DDS
 
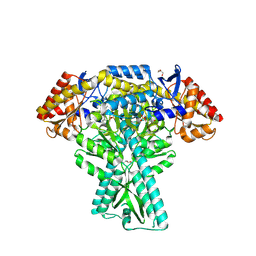 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PLP | | 分子名称: | ACETIC ACID, CrmG, GLYCEROL, ... | | 著者 | Xu, J, Feng, Z, Liu, J. | | 登録日 | 2015-08-25 | | 公開日 | 2016-08-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5DDW
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with the PMP external aldimine adduct with Caerulomycin M | | 分子名称: | CrmG, GLYCEROL, [5-hydroxy-4-({(E)-[(4-hydroxy-2,2'-bipyridin-6-yl)methylidene]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Xu, J, Feng, Z, Liu, J. | | 登録日 | 2015-08-25 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
