1PVZ
 
 | | Solution Structure of BmP07, A Novel Potassium Channel Blocker from Scorpion Buthus martensi Karsch, 15 structures | | 分子名称: | K+ toxin-like peptide | | 著者 | Wu, H, Zhang, N, Wang, Y, Zhang, Q, Ou, L, Li, M, Hu, G. | | 登録日 | 2003-06-29 | | 公開日 | 2004-05-18 | | 最終更新日 | 2018-06-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of BmKK2, a new potassium channel blocker from the venom of chinese scorpion Buthus martensi Karsch
PROTEINS, 55, 2004
|
|
1YW9
 
 | | h-MetAP2 complexed with A849519 | | 分子名称: | 2-[({2-[(1Z)-3-(DIMETHYLAMINO)PROP-1-ENYL]-4-FLUOROPHENYL}SULFONYL)AMINO]-5,6,7,8-TETRAHYDRONAPHTHALENE-1-CARBOXYLIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | 著者 | Park, C.H. | | 登録日 | 2005-02-17 | | 公開日 | 2006-02-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Discovery and optimization of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2: a structural basis for the reduction of albumin binding.
J.Med.Chem., 49, 2006
|
|
2DDY
 
 | | Solution Structure of Matrilysin (MMP-7) Complexed to Constraint Conformational Sulfonamide Inhibitor | | 分子名称: | (1R)-N,6-DIHYDROXY-7-METHOXY-2-[(4-METHOXYPHENYL)SULFONYL]-1,2,3,4-TETRAHYDROISOQUINOLINE-1-CARBOXAMIDE, CALCIUM ION, Matrilysin, ... | | 著者 | Zheng, X.H, Ou, L. | | 登録日 | 2006-02-06 | | 公開日 | 2007-02-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Matrilysin (MMP-7) Complexed to Constraint Conformational Sulfonamide Inhibitor
to be published
|
|
3LPI
 
 | | Structure of BACE Bound to SCH745132 | | 分子名称: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylsulfonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | 著者 | Strickland, C, Cumming, J. | | 登録日 | 2010-02-05 | | 公開日 | 2010-04-14 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LPK
 
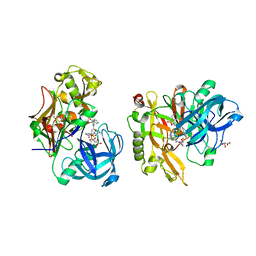 | | Structure of BACE Bound to SCH747123 | | 分子名称: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-{(2R)-4-[(3-methylphenyl)sulfonyl]piperazin-2-yl}ethyl]-3-{[(2R)-2-(methoxymethyl)pyrrolidin-1-yl]carbonyl}-5-methylbenzamide | | 著者 | Strickland, C, Cumming, J. | | 登録日 | 2010-02-05 | | 公開日 | 2010-04-14 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LPJ
 
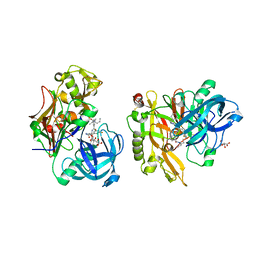 | | Structure of BACE Bound to SCH743641 | | 分子名称: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-[(1S,2S)-2-[(2R)-4-benzylpiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | 著者 | Strickland, C, Cumming, J. | | 登録日 | 2010-02-05 | | 公開日 | 2010-04-14 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6PBC
 
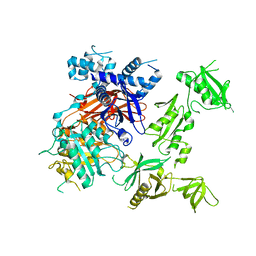 | | Structural basis for the activation of PLC-gamma isozymes by phosphorylation and cancer-associated mutations | | 分子名称: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma,1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, CALCIUM ION, SODIUM ION | | 著者 | Hajicek, N, Sondek, J. | | 登録日 | 2019-06-13 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Structural basis for the activation of PLC-gamma isozymes by phosphorylation and cancer-associated mutations.
Elife, 8, 2019
|
|
3LNK
 
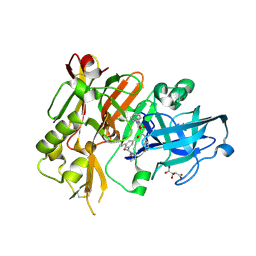 | | Structure of BACE bound to SCH743813 | | 分子名称: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylcarbonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | 著者 | Orth, P, Cumming, J. | | 登録日 | 2010-02-02 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | 著者 | Zhang, X, Chen, Z. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
1YW8
 
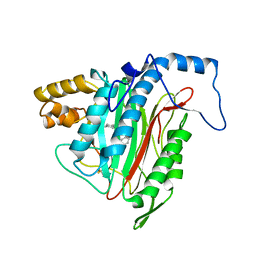 | | h-MetAP2 complexed with A751277 | | 分子名称: | 2-[(PHENYLSULFONYL)AMINO]-5,6,7,8-TETRAHYDRONAPHTHALENE-1-CARBOXYLIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | 著者 | Park, C.H. | | 登録日 | 2005-02-17 | | 公開日 | 2006-02-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Discovery and optimization of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2: a structural basis for the reduction of albumin binding.
J.Med.Chem., 49, 2006
|
|
1YW7
 
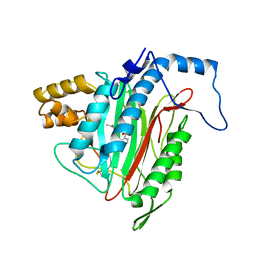 | | h-MetAP2 complexed with A444148 | | 分子名称: | 5-METHYL-2-[(PHENYLSULFONYL)AMINO]BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | 著者 | Park, C.H. | | 登録日 | 2005-02-17 | | 公開日 | 2006-02-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery and optimization of anthranilic acid sulfonamides as inhibitors of methionine aminopeptidase-2: a structural basis for the reduction of albumin binding.
J.Med.Chem., 49, 2006
|
|
3TDA
 
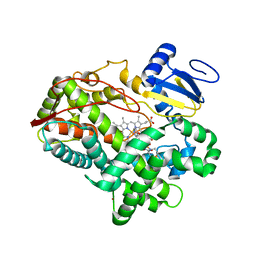 | | Competitive replacement of thioridazine by prinomastat in crystals of cytochrome P450 2D6 | | 分子名称: | Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, Prinomastat, ... | | 著者 | Wang, A, Stout, C.D, Johnson, E.F. | | 登録日 | 2011-08-10 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
3TBG
 
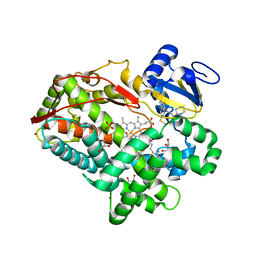 | | Human cytochrome P450 2D6 with two thioridazines bound in active site | | 分子名称: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, Cytochrome P450 2D6, GLYCEROL, ... | | 著者 | Wang, A, Stout, C.D, Johnson, E.F. | | 登録日 | 2011-08-05 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
1M3G
 
 | |
5GSA
 
 | | EED in complex with an allosteric PRC2 inhibitor | | 分子名称: | Histone-lysine N-methyltransferase EZH2, N-(furan-2-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, Polycomb protein EED | | 著者 | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | 登録日 | 2016-08-15 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of
Nat. Chem. Biol., 13, 2017
|
|
6AX3
 
 | | Complex structure of JMJD5 and Symmetric Dimethyl-Arginine (SDMA) | | 分子名称: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, N3, ... | | 著者 | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | 登録日 | 2017-09-06 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
3Q96
 
 | | B-Raf kinase domain in complex with a tetrahydronaphthalene inhibitor | | 分子名称: | (2S)-N-[3-(2-aminopropan-2-yl)-5-(trifluoromethyl)phenyl]-7-[(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-4-yl)oxy]-1,2,3,4-tetrahydronaphthalene-2-carboxamide, Serine/threonine-protein kinase B-raf | | 著者 | Sintchak, M.D, Aertgeerts, K, Yano, J. | | 登録日 | 2011-01-07 | | 公開日 | 2011-03-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Design and optimization of potent and orally bioavailable tetrahydronaphthalene raf inhibitors.
J.Med.Chem., 54, 2011
|
|
6AVS
 
 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | 分子名称: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | 著者 | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | 登録日 | 2017-09-04 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
4N0N
 
 | | Crystal structure of Arterivirus nonstructural protein 10 (helicase) | | 分子名称: | MAGNESIUM ION, Replicase polyprotein 1ab, SULFATE ION, ... | | 著者 | Deng, Z, Chen, Z. | | 登録日 | 2013-10-02 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
8X71
 
 | | Crystal structure of Peroxiredoxin I in complex with compound 19-064 | | 分子名称: | Peroxiredoxin-1, methyl 3-[[(2~{R},4~{a}~{S},6~{a}~{R},6~{a}~{S},14~{a}~{S},14~{b}~{R})-2,4~{a},6~{a},6~{a},9,14~{a}-hexamethyl-10-oxidanyl-11-oxidanylidene-1,3,4,5,6,13,14,14~{b}-octahydropicen-2-yl]carbamoylamino]oxetane-3-carboxylate | | 著者 | Zhang, H, Luo, C. | | 登録日 | 2023-11-22 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Discovery of a Novel Orally Bioavailable FLT3-PROTAC Degrader for Efficient Treatment of Acute Myeloid Leukemia and Overcoming Resistance of FLT3 Inhibitors.
J.Med.Chem., 67, 2024
|
|
8X73
 
 | | Crystal structure of Peroxiredoxin I in complex with compound 19-069 | | 分子名称: | Peroxiredoxin-1, methyl (2~{S})-2-[[(2~{R},4~{a}~{S},6~{a}~{R},6~{a}~{S},14~{a}~{S},14~{b}~{R})-2,4~{a},6~{a},6~{a},9,14~{a}-hexamethyl-10-oxidanyl-11-oxidanylidene-1,3,4,5,6,13,14,14~{b}-octahydropicen-2-yl]carbamoylamino]-3-oxidanyl-propanoate | | 著者 | Zhang, H, Luo, C. | | 登録日 | 2023-11-22 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Discovery of a Novel Orally Bioavailable FLT3-PROTAC Degrader for Efficient Treatment of Acute Myeloid Leukemia and Overcoming Resistance of FLT3 Inhibitors.
J.Med.Chem., 67, 2024
|
|
4HA5
 
 | | Structure of BACE Bound to (S)-3-(5-(2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl)thiophen-3-yl)benzonitrile | | 分子名称: | 3-{5-[(2E,4S)-2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Mandal, M. | | 登録日 | 2012-09-25 | | 公開日 | 2012-10-17 | | 最終更新日 | 2012-11-21 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3J
 
 | | Structure of BACE Bound to 2-fluoro-5-(5-(2-imino-3-methyl-4-oxo-6-phenyloctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-2-yl)benzonitrile | | 分子名称: | 2-fluoro-5-{5-[(2E,4aR,7aR)-2-imino-3-methyl-4-oxo-6-phenyloctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-2-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Mandal, M. | | 登録日 | 2012-09-13 | | 公開日 | 2012-10-17 | | 最終更新日 | 2012-11-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3F
 
 | | Structure of BACE Bound to 3-(5-((7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-1H-pyrrolo[3,4-d]pyrimidin-7a-yl)thiophen-3-yl)benzonitrile | | 分子名称: | 3-{5-[(2E,4aR,7aR)-2-imino-6-(6-methoxypyridin-2-yl)-3-methyl-4-oxooctahydro-7aH-pyrrolo[3,4-d]pyrimidin-7a-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Mandal, M. | | 登録日 | 2012-09-13 | | 公開日 | 2012-11-07 | | 最終更新日 | 2012-11-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3G
 
 | | Structure of BACE Bound to 2-((7aR)-7a-(4-(3-cyanophenyl)thiophen-2-yl)-2-imino-3-methyl-4-oxohexahydro-1H-pyrrolo[3,4-d]pyrimidin-6(2H)-yl)nicotinonitrile | | 分子名称: | 2-{(2E,4aR,7aR)-7a-[4-(3-cyanophenyl)thiophen-2-yl]-2-imino-3-methyl-4-oxooctahydro-6H-pyrrolo[3,4-d]pyrimidin-6-yl}pyridine-3-carbonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Mandal, M. | | 登録日 | 2012-09-13 | | 公開日 | 2012-11-07 | | 最終更新日 | 2012-11-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
