4QPI
 
 | | Crystal structure of hepatitis A virus | | 分子名称: | CHLORIDE ION, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | 登録日 | 2014-06-23 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
8DGB
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Q192T Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-23 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DCZ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) M165Y Mutant in Complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD1
 
 | | SARS-CoV-2 Main Protease (Mpro) H164N Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFN
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H164N Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFE
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD9
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
3CMZ
 
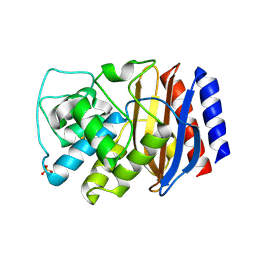 | | TEM-1 Class-A beta-lactamase L201P mutant apo structure | | 分子名称: | Beta-lactamase TEM, PHOSPHATE ION | | 著者 | Marciano, D.C, Wang, X, Wang, J, Chen, Y, Thomas, V.L, Shoichet, B.K, Palzkill, T. | | 登録日 | 2008-03-24 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase
J.Mol.Biol., 384, 2008
|
|
4U7U
 
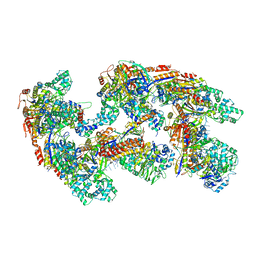 | | Crystal structure of RNA-guided immune Cascade complex from E.coli | | 分子名称: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | 著者 | Zhao, H, Sheng, G, Wang, J, Wang, M, Bunkoczi, G, Gong, W, Wei, Z, Wang, Y. | | 登録日 | 2014-07-31 | | 公開日 | 2014-08-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.003 Å) | | 主引用文献 | Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Nature, 515, 2014
|
|
1IG9
 
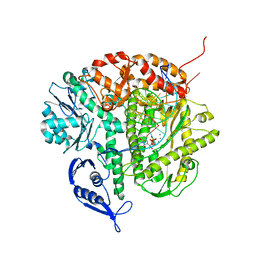 | | Structure of the Replicating Complex of a Pol Alpha Family DNA Polymerase | | 分子名称: | 5'-D(*AP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*(DOC))-3', CALCIUM ION, ... | | 著者 | Franklin, M.C, Wang, J, Steitz, T.A. | | 登録日 | 2001-04-17 | | 公開日 | 2001-06-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of the Replicating Complex of a Pol Alpha Family DNA Polymerase
Cell(Cambridge,Mass.), 105, 2001
|
|
3CD4
 
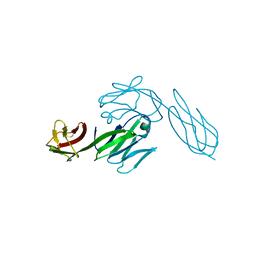 | |
2BM2
 
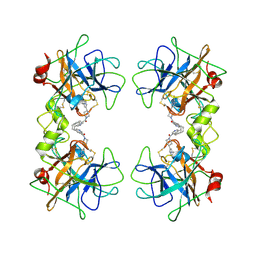 | | human beta-II tryptase in complex with 4-(3-Aminomethyl-phenyl)- piperidin-1-yl-(5-phenethyl- pyridin-3-yl)-methanone | | 分子名称: | 1-[3-(1-{[5-(2-PHENYLETHYL)PYRIDIN-3-YL]CARBONYL}PIPERIDIN-4-YL)PHENYL]METHANAMINE, HUMAN BETA2 TRYPTASE | | 著者 | Maignan, S, Guilloteau, J.-P, Dupuy, A, Levell, J, Astles, P, Eastwood, P, Cairns, J, Houille, O, Aldous, S, Merriman, G, Whiteley, B, Pribish, J, Czekaj, M, Liang, G, Davidson, J, Harrison, T, Morley, A, Watson, S, Fenton, G, Mccarthy, C, Romano, J, Mathew, R, Engers, D, Gardyan, M, Sides, K, Kwong, J, Tsay, J, Rebello, S, Shen, L, Wang, J, Luo, Y, Giardino, O, Lim, H.-K, Smith, K, Pauls, H. | | 登録日 | 2005-03-09 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure Based Design of 4-(3-Aminomethylphenyl) Piperidinyl-1-Amides: Novel, Potent, Selective, and Orally Bioavailable Inhibitors of Bii Tryptase
Bioorg.Med.Chem., 13, 2005
|
|
1JE9
 
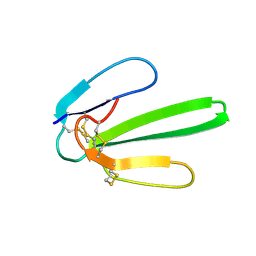 | | NMR SOLUTION STRUCTURE OF NT2 | | 分子名称: | SHORT NEUROTOXIN II | | 著者 | Cheng, Y, Wang, W, Wang, J. | | 登録日 | 2001-06-16 | | 公開日 | 2001-07-04 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function relationship of three neurotoxins from the venom of Naja kaouthia: a comparison between the NMR-derived structure of NT2 with its homologues, NT1 and NT3
BIOCHIM.BIOPHYS.ACTA, 1594, 2002
|
|
3HVT
 
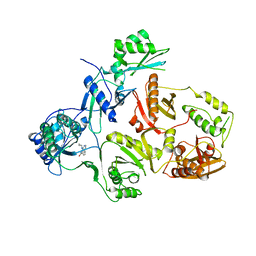 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | 著者 | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | 登録日 | 1994-07-25 | | 公開日 | 1994-10-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
6YG3
 
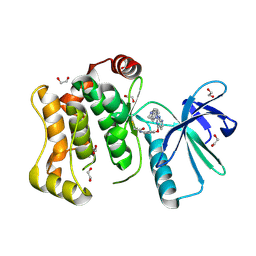 | | Crystal structure of MKK7 (MAP2K7) covalently bound with CPT1-70-1 | | 分子名称: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, N-(4-((2-((4-(4-methylpiperazin-1-yl)phenyl)amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy)phenyl)acrylamide | | 著者 | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2020-03-27 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
6YG7
 
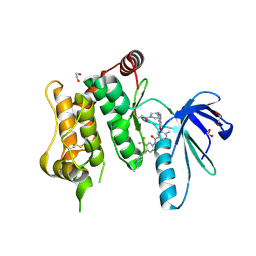 | | Crystal structure of MKK7 (MAP2K7) covalently bound with type-II inhibitor SB1-G-23 | | 分子名称: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-[[(3~{R})-1-propanoylpyrrolidin-3-yl]amino]pyrimidin-4-yl]oxy-benzamide | | 著者 | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2020-03-27 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
6YG6
 
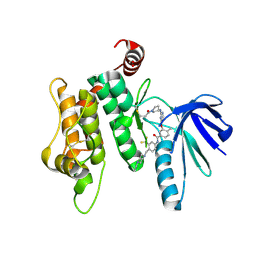 | | Crystal structure of MKK7 (MAP2K7) covalently bound with type-II inhibitor TL10-105 | | 分子名称: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-[[(3~{S})-1-propanoylpyrrolidin-3-yl]amino]pyrimidin-4-yl]oxy-benzamide | | 著者 | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2020-03-27 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
8H2H
 
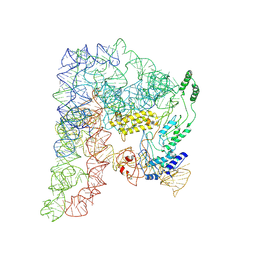 | | Cryo-EM structure of a Group II Intron Complexed with its Reverse Transcriptase | | 分子名称: | Group II intron-encoded protein LtrA, LtrB, RNA (5'-R(P*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*C)-3') | | 著者 | Liu, N, Dong, X.L, Qu, G.S, Wang, J, Wang, H.W, Belfort, M. | | 登録日 | 2022-10-06 | | 公開日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Functionalized graphene grids with various charges for single-particle cryo-EM.
Nat Commun, 13, 2022
|
|
1IH7
 
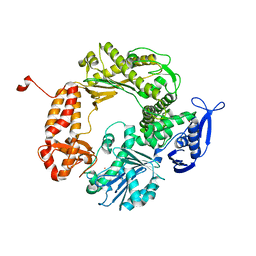 | | High-Resolution Structure of Apo RB69 DNA Polymerase | | 分子名称: | DNA POLYMERASE, GUANOSINE, POTASSIUM ION | | 著者 | Franklin, M.C, Wang, J, Steitz, T.A. | | 登録日 | 2001-04-18 | | 公開日 | 2001-06-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structure of the Replicating Complex of a Pol alpha Family DNA Polymerase
Cell(Cambridge,Mass.), 105, 2001
|
|
6WTT
 
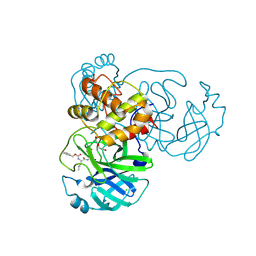 | | Crystals Structure of the SARS-CoV-2 (COVID-19) main protease with inhibitor GC-376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Sacco, M, Ma, C, Chen, Y, Wang, J. | | 登録日 | 2020-05-03 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease.
Cell Res., 30, 2020
|
|
2EFG
 
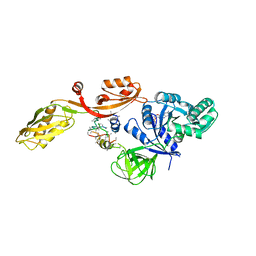 | | TRANSLATIONAL ELONGATION FACTOR G COMPLEXED WITH GDP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, PROTEIN (ELONGATION FACTOR G DOMAIN 3), PROTEIN (ELONGATION FACTOR G) | | 著者 | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | 登録日 | 1998-09-23 | | 公開日 | 1999-09-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
7YVR
 
 | | Crystal Structure of L-Threonine Aldolase from Neptunomonas marina | | 分子名称: | GLYCEROL, L-threonine aldolase | | 著者 | He, Y.Z, Wang, J, Yan, W.P, Zhang, Y, Feng, Y. | | 登録日 | 2022-08-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery and Engineering of the L-Threonine Aldolase from Neptunomonas marine for the Efficient Synthesis of beta-Hydroxy-alpha-amino Acids via C-C Formation
Acs Catalysis, 2023
|
|
8D4L
 
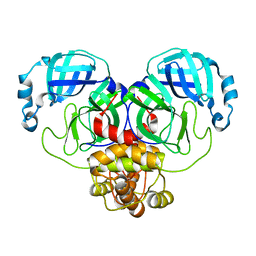 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4K
 
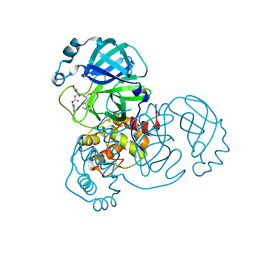 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4M
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
