6IQI
 
 | | crystal structure of Arabidopsis thaliana Profilin 2 | | 分子名称: | Profilin-2 | | 著者 | Qiao, Z, Gao, Y. | | 登録日 | 2018-11-08 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and computational examination of theArabidopsisprofilin-Poly-P complex reveals mechanistic details in profilin-regulated actin assembly.
J.Biol.Chem., 294, 2019
|
|
6IQF
 
 | |
6IQJ
 
 | |
8H5U
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-021 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb-021, ... | | 著者 | Yang, J, Lin, S, Lu, G.W. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Development of a bispecific nanobody conjugate broadly neutralizes diverse SARS-CoV-2 variants and structural basis for its broad neutralization.
Plos Pathog., 19, 2023
|
|
8H5T
 
 | |
6J67
 
 | |
6IQK
 
 | | crystal structure of Arabidopsis thaliana Profilin 3 | | 分子名称: | AtPRF3, Profilin-5 | | 著者 | Qiao, Z, Gao, Y. | | 登録日 | 2018-11-08 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural and computational examination of theArabidopsisprofilin-Poly-P complex reveals mechanistic details in profilin-regulated actin assembly.
J.Biol.Chem., 294, 2019
|
|
6KKN
 
 | | Crystal structure of RuBisCO accumulation factor Raf1 from Anabaena sp. PCC 7120 | | 分子名称: | All5250 protein | | 著者 | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | 登録日 | 2019-07-26 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
6LRR
 
 | | Cryo-EM structure of RuBisCO-Raf1 from Anabaena sp. PCC 7120 | | 分子名称: | All5250 protein, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | 著者 | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | 登録日 | 2020-01-16 | | 公開日 | 2020-05-13 | | 最終更新日 | 2020-07-01 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
6LRC
 
 | | Human cGAS catalytic domain bound with the inhibitor PF-06928215 | | 分子名称: | (1R,2S)-2-[(7-hydroxy-5-phenylpyrazolo[1,5-a]pyrimidine-3-carbonyl)amino]cyclohexane-1-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | 著者 | Zhao, W.F, Xu, Y.C. | | 登録日 | 2020-01-15 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.831 Å) | | 主引用文献 | In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing.
J.Chem.Inf.Model., 60, 2020
|
|
6NZR
 
 | |
7X4I
 
 | |
8H2T
 
 | | Cryo-EM structure of IadD/E dioxygenase bound with IAA | | 分子名称: | 1H-INDOL-3-YLACETIC ACID, Aromatic-ring-hydroxylating dioxygenase beta subunit, FE (III) ION, ... | | 著者 | Yu, G, Li, Z, Zhang, H. | | 登録日 | 2022-10-07 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Structural and biochemical characterization of the key components of an auxin degradation operon from the rhizosphere bacterium Variovorax.
Plos Biol., 21, 2023
|
|
2NWM
 
 | |
2QN1
 
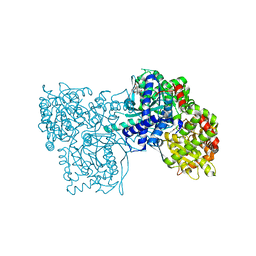 | | Glycogen Phosphorylase b in complex with asiatic acid | | 分子名称: | Glycogen phosphorylase, muscle form, asiatic acid | | 著者 | Zographos, S.E, Leonidas, D.D, Alexacou, K.-M, Hayes, J, Oikonomakos, N.G. | | 登録日 | 2007-07-17 | | 公開日 | 2008-06-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Naturally occurring pentacyclic triterpenes as inhibitors of glycogen phosphorylase: synthesis, structure-activity relationships, and X-ray crystallographic studies
J.Med.Chem., 51, 2008
|
|
2QN2
 
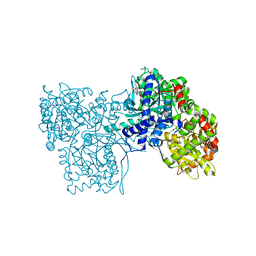 | | Glycogen Phosphorylase b in complex with Maslinic Acid | | 分子名称: | Glycogen phosphorylase, muscle form, maslinic acid | | 著者 | Zographos, S.E, Leonidas, D.D, Alexacou, K.-M, Hayes, J, Oikonomakos, N.G. | | 登録日 | 2007-07-17 | | 公開日 | 2008-06-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.698 Å) | | 主引用文献 | Naturally occurring pentacyclic triterpenes as inhibitors of glycogen phosphorylase: synthesis, structure-activity relationships, and X-ray crystallographic studies
J.Med.Chem., 51, 2008
|
|
5Y8V
 
 | | Crystal structure of GAS41 | | 分子名称: | MAGNESIUM ION, YEATS domain-containing protein 4 | | 著者 | Wang, Y, Hao, Q. | | 登録日 | 2017-08-21 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8IA3
 
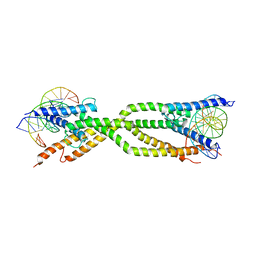 | | Crystal structure of human USF2 bHLHLZ domain in complex with DNA | | 分子名称: | DNA (5'-D(*GP*CP*GP*CP*GP*TP*CP*AP*CP*GP*TP*GP*CP*CP*CP*GP*TP*C)-3'), DNA (5'-D(P*GP*AP*CP*GP*GP*GP*CP*AP*CP*GP*TP*GP*AP*CP*GP*CP*GP*C)-3'), Upstream stimulatory factor 2 | | 著者 | Huang, C, Fang, P, Wang, J. | | 登録日 | 2023-02-07 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Tetramerization of upstream stimulating factor USF2 requires the elongated bent leucine zipper of the bHLH-LZ domain.
J.Biol.Chem., 299, 2023
|
|
7XE4
 
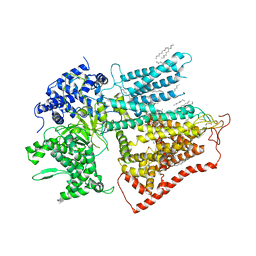 | | structure of a membrane-bound glycosyltransferase | | 分子名称: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | 登録日 | 2022-03-29 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | 分子名称: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPF
 
 | |
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | 登録日 | 2020-12-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
