4LIQ
 
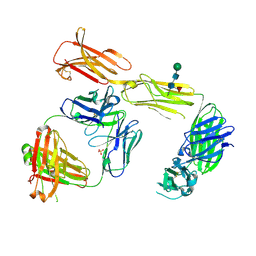 | | Structure of the extracellular domain of human CSF-1 receptor in complex with the Fab fragment of RG7155 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment RG7155 heavy chain, Fab fragment RG7155 light chain, ... | | 著者 | Benz, J, Gorr, I.H, Hertenberger, H, Ries, C.H. | | 登録日 | 2013-07-03 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy
Cancer Cell, 25, 2014
|
|
8E5N
 
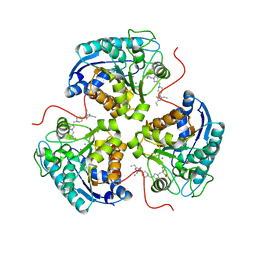 | | Structure of ARG1 complex with pyrrolidine-based non-boronic acid inhibitor 10 | | 分子名称: | 1-{[(3S,4S)-3-(3-fluorophenyl)-4-{[4-(1,3,4-triethyl-1H-pyrazol-5-yl)piperidin-1-yl]methyl}pyrrolidin-1-yl]methyl}cyclopentane-1-carboxylic acid, Arginase-1, MANGANESE (II) ION | | 著者 | Palte, R.L, Gathiaka, S. | | 登録日 | 2022-08-22 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.538 Å) | | 主引用文献 | Discovery of non-boronic acid Arginase 1 inhibitors through virtual screening and biophysical methods.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
8E5M
 
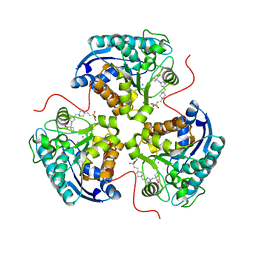 | | Structure of ARG1 complex with pyrrolidine-based non-boronic acid inhibitor 6 | | 分子名称: | 1-{[(3S,4S)-3-({4-[2-(4-fluorobenzene-1-sulfonyl)ethyl]piperidin-1-yl}methyl)-4-(3-fluorophenyl)pyrrolidin-1-yl]methyl}cyclopentane-1-carboxylic acid, Arginase-1, MANGANESE (II) ION | | 著者 | Palte, R.L, Gathiaka, S. | | 登録日 | 2022-08-22 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Discovery of non-boronic acid Arginase 1 inhibitors through virtual screening and biophysical methods.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
1OBH
 
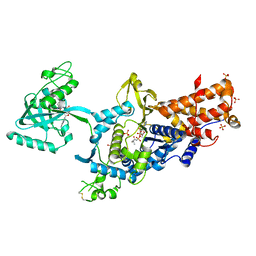 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A PRE-TRANSFER EDITING SUBSTRATE ANALOGUE IN BOTH SYNTHETIC ACTIVE SITE AND EDITING SITE | | 分子名称: | LEUCYL-TRNA SYNTHETASE, MERCURY (II) ION, NORVALINE, ... | | 著者 | Cusack, S, Yaremchuk, A, Tukalo, M. | | 登録日 | 2003-01-31 | | 公開日 | 2003-05-09 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and Mechanistic Basis of Pre- and Posttransfer Editing by Leucyl-tRNA Synthetase
Mol.Cell, 11, 2003
|
|
1OBC
 
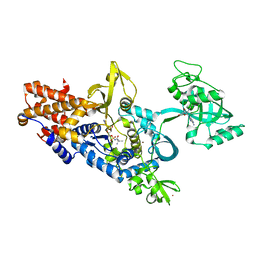 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A POST-TRANSFER EDITING SUBSTRATE ANALOGUE | | 分子名称: | 2'-AMINO-2'-DEOXYADENOSINE, LEUCINE, LEUCYL-TRNA SYNTHETASE, ... | | 著者 | Cusack, S, Yaremchuk, A, Tukalo, M. | | 登録日 | 2003-01-30 | | 公開日 | 2003-05-09 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and Mechanistic Basis of Pre- and Posttransfer Editing by Leucyl-tRNA Synthetase
Mol.Cell, 11, 2003
|
|
5LL9
 
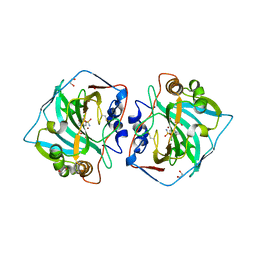 | | Crystal structure of human carbonic anhydrase isozyme XII with 4-(1H-benzimidazol-1-ylacetyl)-2-chlorobenzenesulfonamide | | 分子名称: | 1,2-ETHANEDIOL, 4-[2-(benzimidazol-1-yl)ethanoyl]-2-chloranyl-benzenesulfonamide, Carbonic anhydrase 12, ... | | 著者 | Smirnov, A, Manakova, E, Grazulis, S. | | 登録日 | 2016-07-27 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Combinatorial Design of Isoform-Selective N-Alkylated Benzimidazole-Based Inhibitors of Carbonic Anhydrases
Chemistryselect, 2017
|
|
2P3T
 
 | |
6E62
 
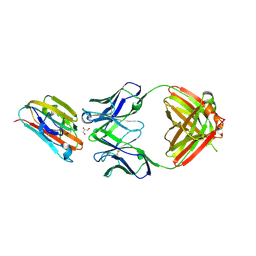 | | Crystal structure of malaria transmission-blocking antigen Pfs48/45 6C in complex with antibody 85RF45.1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 85RF45.1 Fab heavy chain, 85RF45.1 Fab light chain, ... | | 著者 | Kundu, P, Semesi, A, Julien, J.P. | | 登録日 | 2018-07-23 | | 公開日 | 2018-11-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural delineation of potent transmission-blocking epitope I on malaria antigen Pfs48/45.
Nat Commun, 9, 2018
|
|
6E63
 
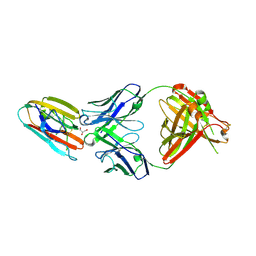 | | Crystal structure of malaria transmission-blocking antigen Pfs48/45 6C in complex with antibody TB31F | | 分子名称: | GLYCEROL, Pf48/45, TB31F Fab heavy chain, ... | | 著者 | Kundu, P, Semesi, A, Julien, J.P. | | 登録日 | 2018-07-23 | | 公開日 | 2018-11-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural delineation of potent transmission-blocking epitope I on malaria antigen Pfs48/45.
Nat Commun, 9, 2018
|
|
6E65
 
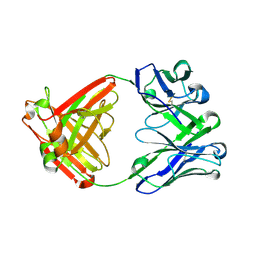 | |
6E64
 
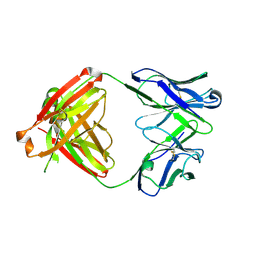 | |
7B3E
 
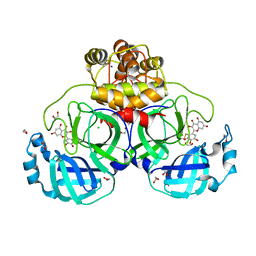 | | Crystal structure of myricetin covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2020-11-30 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen.
Acs Pharmacol Transl Sci, 4, 2021
|
|
6EDS
 
 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Glucagon and Substrate-selective Macrocyclic Inhibitor 63 | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Tan, G.A, Seeliger, M.A, Maianti, J.P, Liu, D.R, Welsh, A.J. | | 登録日 | 2018-08-10 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.18071723 Å) | | 主引用文献 | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
4MM3
 
 | |
4L8S
 
 | |
4L9L
 
 | |
4LCC
 
 | | Crystal structure of a human MAIT TCR in complex with a bacterial antigen bound to humanized bovine MR1 | | 分子名称: | 1-deoxy-1-[6-(hydroxymethyl)-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl]-D-arabinitol, Beta-2-microglobulin, MHC class I-related protein, ... | | 著者 | Lopez-Sagaseta, J, Adams, E.J. | | 登録日 | 2013-06-21 | | 公開日 | 2013-10-16 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.263 Å) | | 主引用文献 | MAIT Recognition of a Stimulatory Bacterial Antigen Bound to MR1.
J.Immunol., 191, 2013
|
|
7ALI
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup P2(1)). | | 分子名称: | 3C-like proteinase | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | 登録日 | 2020-10-06 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7ALH
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup C2). | | 分子名称: | 3C-like proteinase | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | 登録日 | 2020-10-06 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
1UMN
 
 | | Crystal structure of Dps-like peroxide resistance protein (Dpr) from Streptococcus suis | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Kauko, A, Haataja, S, Pulliainen, A, Finne, J, Papageorgiou, A.C. | | 登録日 | 2003-08-26 | | 公開日 | 2004-04-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of Streptococcus suis Dps-like peroxide resistance protein Dpr: implications for iron incorporation.
J. Mol. Biol., 338, 2004
|
|
4OJQ
 
 | |
4OKS
 
 | |
3ZJ6
 
 | | Crystal of Raucaffricine Glucosidase in complex with inhibitor | | 分子名称: | (1R,2S,3S,4R,5R)-4-(cyclohexylmethylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | 著者 | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | 登録日 | 2013-01-17 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
3ZJ8
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-2 | | 分子名称: | (1R,2S,3S,4R,5R)-4-[(4-bromophenyl)methylamino]-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | 著者 | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | 登録日 | 2013-01-17 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
3ZJ7
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-1 | | 分子名称: | (1R,2S,3S,4R,5R)-4-(cyclohexylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | 著者 | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | 登録日 | 2013-01-17 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
