6EOR
 
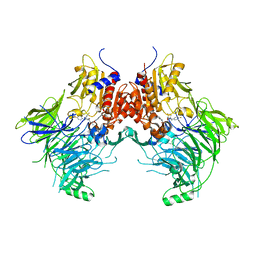 | | DPP9 - 1G244 | | 分子名称: | (2~{S})-2-azanyl-4-[4-[bis(4-fluorophenyl)methyl]piperazin-1-yl]-1-(1,3-dihydroisoindol-2-yl)butane-1,4-dione, Dipeptidyl peptidase 9 | | 著者 | Ross, B.R, Huber, R. | | 登録日 | 2017-10-10 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EOQ
 
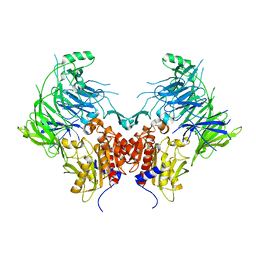 | | DPP9 - Apo | | 分子名称: | Dipeptidyl peptidase 9 | | 著者 | Ross, B.R, Huber, R. | | 登録日 | 2017-10-10 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8E4R
 
 | |
8E4J
 
 | |
8QV7
 
 | |
6SQ0
 
 | |
2PJW
 
 | |
6SUO
 
 | | ERa_L536S (L536S/C381S/C471S,C530S) in complex with a tricyclic indole (compound 6) | | 分子名称: | (~{E})-3-[3,5-bis(fluoranyl)-4-[(1~{R},3~{R})-2-(2-fluoranyl-2-methyl-propyl)-1,3-dimethyl-4,9-dihydro-3~{H}-pyrido[3,4-b]indol-1-yl]phenyl]prop-2-enoic acid, Estrogen receptor | | 著者 | Breed, J. | | 登録日 | 2019-09-16 | | 公開日 | 2019-10-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Building Bridges in a Series of Estrogen Receptor Degraders: An Application of Metathesis in Medicinal Chemistry.
Acs Med.Chem.Lett., 10, 2019
|
|
8VU0
 
 | |
6XGW
 
 | | ISCth4 transposase, pre-reaction complex, PRC | | 分子名称: | DNA (32-MER), Mutator family transposase | | 著者 | Kosek, D, Dyda, F. | | 登録日 | 2020-06-18 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.
Embo J., 40, 2021
|
|
8R5R
 
 | | Structure of apo TDO with a bound inhibitor | | 分子名称: | 3-chloranyl-~{N}-[(1~{S})-1-(6-chloranylpyridin-3-yl)-2-phenyl-ethyl]aniline, Tryptophan 2,3-dioxygenase, alpha-methyl-L-tryptophan | | 著者 | Wicki, M, Mac Sweeney, A. | | 登録日 | 2023-11-17 | | 公開日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.078 Å) | | 主引用文献 | Discovery and binding mode of a small molecule inhibitor of the apo form of human TDO2
Biorxiv, 2024
|
|
8R5Q
 
 | | Structure of apo TDO with a bound inhibitor | | 分子名称: | 3-chloranyl-~{N}-[(1~{S})-1-(6-chloranylpyridin-3-yl)-2-phenyl-ethyl]aniline, Tryptophan 2,3-dioxygenase, alpha-methyl-L-tryptophan | | 著者 | Wicki, M, Mac Sweeney, A. | | 登録日 | 2023-11-17 | | 公開日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Discovery and binding mode of a small molecule inhibitor of the apo form of human TDO2
Biorxiv, 2024
|
|
8R12
 
 | | Structure of compound 8 bound to SARS-CoV-2 main protease | | 分子名称: | 2-[[4-(5-chloranylpyridin-3-yl)carbonyl-1,4-diazepan-1-yl]methyl]benzenecarbonitrile, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.587 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R11
 
 | | Structure of compound 7 bound to SARS-CoV-2 main protease | | 分子名称: | 1,2-ETHANEDIOL, 1-[(2~{S})-2-(3-chlorophenyl)pyrrolidin-1-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R14
 
 | | Structure of compound 11 bound to SARS-CoV-2 main protease | | 分子名称: | (5-chloranylpyridin-3-yl)-[4-[(2-chlorophenyl)methyl]-1,4-diazepan-1-yl]methanone, 3C-like proteinase, BROMIDE ION, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.336 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8R16
 
 | | Structure of compound 12 bound to SARS-CoV-2 main protease | | 分子名称: | 1,2-ETHANEDIOL, 1-[6,7-bis(chloranyl)-3,4-dihydro-1H-isoquinolin-2-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | 著者 | Mac Sweeney, A, Hazemann, J. | | 登録日 | 2023-11-01 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
2CAR
 
 | | Crystal Structure Of Human Inosine Triphosphatase | | 分子名称: | INOSINE TRIPHOSPHATE PYROPHOSPHATASE | | 著者 | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Hogbom, M, Holmberg Schiavone, L, Kotenyova, T, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Nordlund, P. | | 登録日 | 2005-12-22 | | 公開日 | 2006-01-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Crystal Structure of Human Inosine Triphosphatase. Substrate Binding and Implication of the Inosine Triphosphatase Deficiency Mutation P32T.
J.Biol.Chem., 282, 2007
|
|
8EQN
 
 | |
7MU6
 
 | | Ask1 bound to compound 28 | | 分子名称: | 2-methoxy-N-(6-{4-[(2S)-1,1,1-trifluoropropan-2-yl]-4H-1,2,4-triazol-3-yl}pyridin-2-yl)pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase 5 | | 著者 | Chodaparambil, J.V, Marcotte, D.J. | | 登録日 | 2021-05-14 | | 公開日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.165 Å) | | 主引用文献 | Discovery of Potent, Selective, and Brain-Penetrant Apoptosis Signal-Regulating Kinase 1 (ASK1) Inhibitors that Modulate Brain Inflammation In Vivo.
J.Med.Chem., 64, 2021
|
|
2ODC
 
 | |
2ODG
 
 | |
8V8E
 
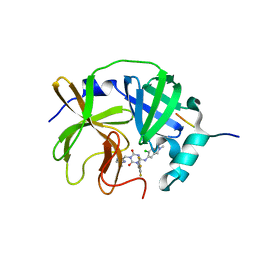 | | Room-temperature X-ray structure of SARS-CoV-2 main protease catalytic domain (residues 1-199-6H) in complex with ensitrelvir (ESV) | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, peptide | | 著者 | Kovalevsky, A, Coates, L. | | 登録日 | 2023-12-05 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Visualizing the Active Site Oxyanion Loop Transition Upon Ensitrelvir Binding and Transient Dimerization of SARS-CoV-2 Main Protease.
J.Mol.Biol., 436, 2024
|
|
8V7W
 
 | |
8V8G
 
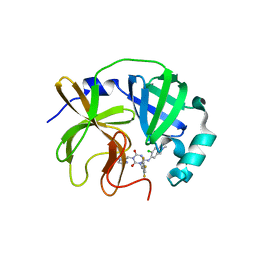 | | Room-temperature X-ray structure of SARS-CoV-2 main protease catalytic domain (residues 1-196) in complex with ensitrelvir (ESV) | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Kovalevsky, A, Coates, L. | | 登録日 | 2023-12-05 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Visualizing the Active Site Oxyanion Loop Transition Upon Ensitrelvir Binding and Transient Dimerization of SARS-CoV-2 Main Protease.
J.Mol.Biol., 436, 2024
|
|
8V7T
 
 | |
