8HI8
 
 | |
8HCI
 
 | |
8HI7
 
 | |
6GWP
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-stab) in Complex with Two Inhibitory Nanobodies (VHH-2g-42, VHH-2w-64) | | 分子名称: | Plasminogen Activator Inhibitor-1, VHH-2g-42, VHH-2w-64 | | 著者 | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | 登録日 | 2018-06-25 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Molecular mechanism of two nanobodies that inhibit PAI-1 activity reveals a modulation at distinct stages of the PAI-1/plasminogen activator interaction.
J.Thromb.Haemost., 18, 2020
|
|
6GWN
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) in Complex with Two Inhibitory Nanobodies (VHH-2g-42, VHH-2w-64) | | 分子名称: | Plasminogen activator inhibitor 1, VHH-2g-42, VHH-2w-64 | | 著者 | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | 登録日 | 2018-06-25 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Molecular mechanism of two nanobodies that inhibit PAI-1 activity reveals a modulation at distinct stages of the PAI-1/plasminogen activator interaction.
J.Thromb.Haemost., 18, 2020
|
|
7VR8
 
 | | Inward-facing structure of human EAAT2 in the substrate-free state | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | 著者 | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2021-10-22 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
7VR7
 
 | | Inward-facing structure of human EAAT2 in the WAY213613-bound state | | 分子名称: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2021-10-22 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
5B0D
 
 | | Polyketide cyclase OAC from Cannabis sativa, Y27W mutant | | 分子名称: | Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B09
 
 | | Polyketide cyclase OAC from Cannabis sativa bound with Olivetolic acid | | 分子名称: | 2,4-bis(oxidanyl)-6-pentyl-benzoic acid, Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0G
 
 | | Polyketide cyclase OAC from Cannabis sativa, H78S mutant | | 分子名称: | Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0B
 
 | | Polyketide cyclase OAC from Cannabis sativa, I7F mutant | | 分子名称: | ACETATE ION, Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B08
 
 | | Polyketide cyclase OAC from Cannabis sativa | | 分子名称: | Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.325 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0A
 
 | | Polyketide cyclase OAC from Cannabis sativa, H5Q mutant | | 分子名称: | Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0E
 
 | | Polyketide cyclase OAC from Cannabis sativa, V59M mutant | | 分子名称: | GLYCEROL, Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.603 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0F
 
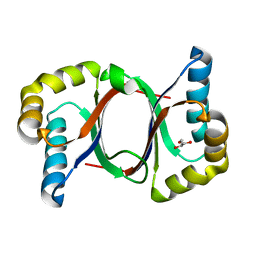 | | Polyketide cyclase OAC from Cannabis sativa, Y72F mutant | | 分子名称: | GLYCEROL, Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0C
 
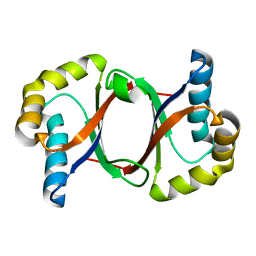 | | Polyketide cyclase OAC from Cannabis sativa, Y27F mutant | | 分子名称: | Olivetolic acid cyclase | | 著者 | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | 登録日 | 2015-10-28 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
8HPM
 
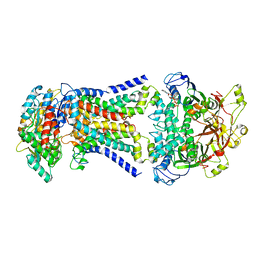 | | LpqY-SugABC in state 2 | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | 登録日 | 2022-12-12 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPN
 
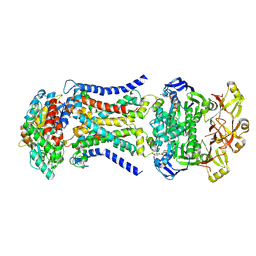 | | LpqY-SugABC in state 3 | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | 登録日 | 2022-12-12 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (4.55 Å) | | 主引用文献 | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPS
 
 | | LpqY-SugABC in state 5 | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | 登録日 | 2022-12-12 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.51 Å) | | 主引用文献 | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPL
 
 | | LpqY-SugABC in state 1 | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | 登録日 | 2022-12-12 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPR
 
 | | LpqY-SugABC in state 4 | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | 登録日 | 2022-12-12 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
5GVT
 
 | |
5YGI
 
 | |
5ZZ3
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 | | 分子名称: | Butyrophilin, subfamily 3, member A3 isoform b variant | | 著者 | Yang, Y.Y, Li, X, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2018-05-30 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
5X5L
 
 | |
