6ZGC
 
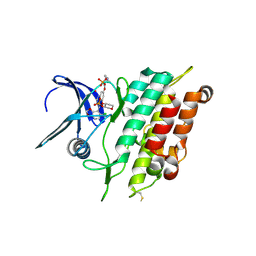 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Saracatinib (AZD0530) | | 分子名称: | Activin receptor type I, N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE, PHOSPHATE ION, ... | | 著者 | Williams, E.P, Galan Bartual, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | 登録日 | 2020-06-18 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Saracatinib is an efficacious clinical candidate for fibrodysplasia ossificans progressiva.
JCI Insight, 6, 2021
|
|
6AGF
 
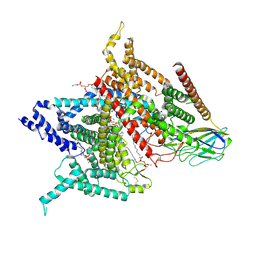 | | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta1 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, X.J, li, Z.Q, Zhou, Q, Shen, H.Z, Wu, K, Huang, X.S, Chen, J.F, Zhang, J.R, Zhu, X.C, Lei, J.L, Xiong, W, Gong, H.P, Xiao, B.L, Yan, N. | | 登録日 | 2018-08-11 | | 公開日 | 2018-10-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta 1.
Science, 362, 2018
|
|
8HIF
 
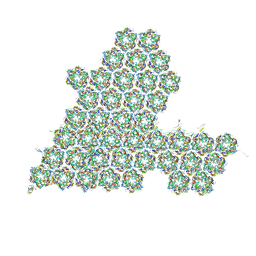 | | One asymmetric unit of Singapore grouper iridovirus capsid | | 分子名称: | Major capsid protein, Penton protein (VP14), VP137, ... | | 著者 | Zhao, Z.N, Liu, C.C, Zhu, D.J, Qi, J.X, Zhang, X.Z, Gao, G.F. | | 登録日 | 2022-11-20 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Near-atomic architecture of Singapore grouper iridovirus and implications for giant virus assembly.
Nat Commun, 14, 2023
|
|
5KQZ
 
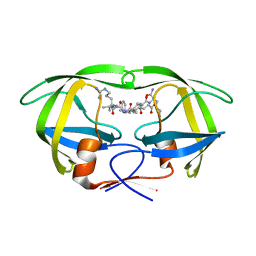 | | Protease E35D-CaP2 | | 分子名称: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, Protease | | 著者 | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | 登録日 | 2016-07-06 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KKN
 
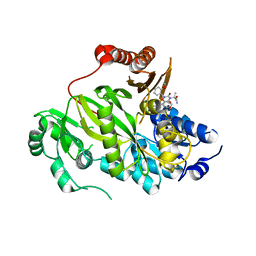 | | Crystal structure of human ACC2 BC domain in complex with ND-646, the primary amide of ND-630 | | 分子名称: | 2-[1-[(2~{R})-2-(2-methoxyphenyl)-2-(oxan-4-yloxy)ethyl]-5-methyl-6-(1,3-oxazol-2-yl)-2,4-bis(oxidanylidene)thieno[2,3-d]pyrimidin-3-yl]-2-methyl-propanamide, Acetyl-CoA carboxylase 2 | | 著者 | Wang, R, Paul, D, Tong, L. | | 登録日 | 2016-06-22 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KQY
 
 | | Protease E35D-DRV | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease E35D-DRV | | 著者 | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | 登録日 | 2016-07-06 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KR1
 
 | | Protease PR5-DRV | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease PR5-DRV | | 著者 | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | 登録日 | 2016-07-06 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KQX
 
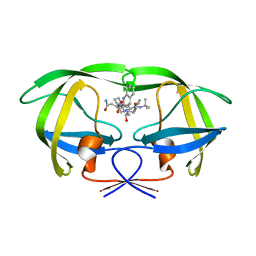 | | Protease E35D-SQV | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease E35D-SQV | | 著者 | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | 登録日 | 2016-07-06 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KZI
 
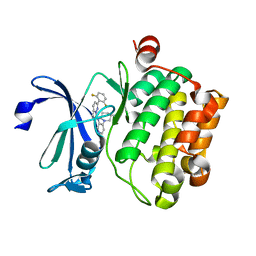 | |
5KR2
 
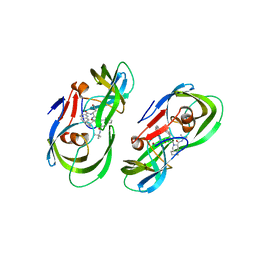 | | Protease PR5-SQV | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease PR5-SQV | | 著者 | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | 登録日 | 2016-07-06 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KR0
 
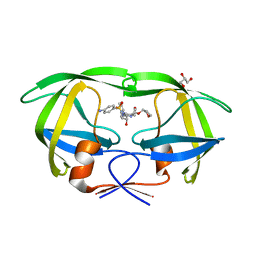 | | Protease E35D-APV | | 分子名称: | GLYCEROL, Protease E35D-APV, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | 著者 | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | 登録日 | 2016-07-06 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
6UTS
 
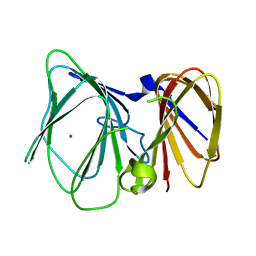 | |
6USY
 
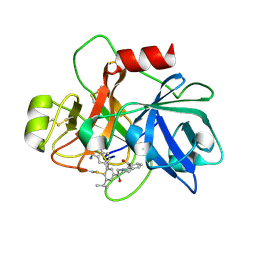 | | COAGULATION FACTOR XI CATALYTIC DOMAIN (C123S) IN COMPLEX WITH NVP-XIV936 | | 分子名称: | 1-[(2S)-2-{3-[(3S)-3-amino-2,3-dihydro-1-benzofuran-5-yl]-5-(propan-2-yl)phenyl}-2-hydroxyethyl]-1H-indole-7-carboxylic acid, Coagulation factor XIa light chain | | 著者 | Weihofen, W.A, Clark, K, Nunes, S. | | 登録日 | 2019-10-28 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
7E3O
 
 | |
7DTD
 
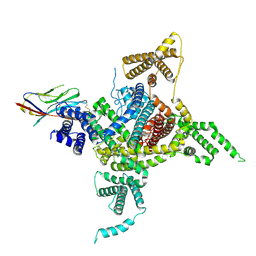 | | Voltage-gated sodium channel Nav1.1 and beta4 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 1 subunit alpha, ... | | 著者 | Yan, N, Pan, X, Li, Z, Huang, G. | | 登録日 | 2021-01-04 | | 公開日 | 2021-04-07 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Comparative structural analysis of human Na v 1.1 and Na v 1.5 reveals mutational hotspots for sodium channelopathies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.764 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4JKV
 
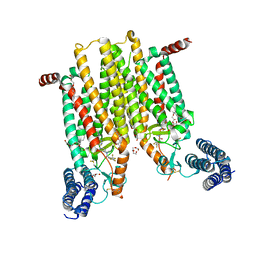 | | Structure of the human smoothened 7TM receptor in complex with an antitumor agent | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-fluoro-N-methyl-N-{1-[4-(1-methyl-1H-pyrazol-5-yl)phthalazin-1-yl]piperidin-4-yl}-2-(trifluoromethyl)benzamide, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wang, C, Wu, H, Katritch, V, Han, G.W, Huang, X, Liu, W, Siu, F.Y, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | 登録日 | 2013-03-11 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structure of the human smoothened receptor bound to an antitumour agent.
Nature, 497, 2013
|
|
4IB4
 
 | | Crystal structure of the chimeric protein of 5-HT2B-BRIL in complex with ergotamine | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, ... | | 著者 | Wacker, D, Wang, C, Katritch, V, Han, G.W, Huang, X, Vardy, E, McCorvy, J.D, Jiang, Y, Chu, M, Siu, F.Y, Liu, W, Xu, H.E, Cherezov, V, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | 登録日 | 2012-12-07 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural features for functional selectivity at serotonin receptors.
Science, 340, 2013
|
|
3DA6
 
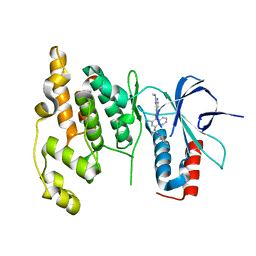 | | Crystal Structure of human JNK3 complexed with N-(3-methyl-4-(3-(2-(methylamino)pyrimidin-4-yl)pyridin-2-yloxy)naphthalen-1-yl)-1H-benzo[d]imidazol-2-amine | | 分子名称: | Mitogen-activated protein kinase 10, N-[3-methyl-4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-1H-benzimidazol-2-amine | | 著者 | Cee, V.J, Cheng, A.C, Romero, K, Bellon, S, Mohr, C, Whittington, D.A, Bready, J, Caenepeel, S, Coxon, A, Deak, H.L, Hodous, B.L, Kim, J.L, Lin, J, Nguyen, H, Olivieri, P.R, Patel, V.F, Wang, L, Hughes, P, Geuns-Meyer, S. | | 登録日 | 2008-05-28 | | 公開日 | 2009-01-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EFW
 
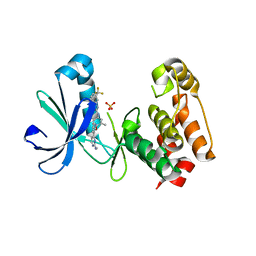 | | Structure of AuroraA with pyridyl-pyrimidine urea inhibitor | | 分子名称: | 1-[3-methyl-4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)phenyl]-3-[3-(trifluoromethyl)phenyl]urea, SULFATE ION, Serine/threonine-protein kinase 6 | | 著者 | Bellon, S.F, Cee, V, Hughes, P, Geuns-Meyer, S, Whittington, D. | | 登録日 | 2008-09-10 | | 公開日 | 2008-12-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3F7O
 
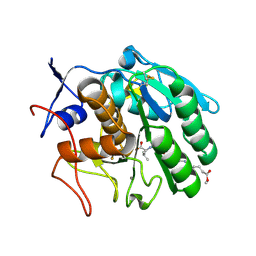 | | Crystal structure of Cuticle-Degrading Protease from Paecilomyces lilacinus (PL646) | | 分子名称: | (MSU)(ALA)(ALA)(PRO)(VAL), CALCIUM ION, Serine protease | | 著者 | Liang, L, Lou, Z, Meng, Z, Rao, Z, Zhang, K. | | 登録日 | 2008-11-10 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
3EWH
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyridyl-pyrimidine benzimidazole inhibitor | | 分子名称: | 1,2-ETHANEDIOL, N-[4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-6-(trifluoromethyl)-1H-benzimidazol-2-amine, vascular endothelial growth factor receptor 2 | | 著者 | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | 登録日 | 2008-10-15 | | 公開日 | 2009-08-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3F7M
 
 | | Crystal structure of apo Cuticle-Degrading Protease (ver112) from Verticillium psalliotae | | 分子名称: | Alkaline serine protease ver112 | | 著者 | Liang, L, Lou, Z, Ye, F, Meng, Z, Rao, Z, Zhang, K. | | 登録日 | 2008-11-09 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
5GHY
 
 | |
8DKF
 
 | |
