2ITK
 
 | | human Pin1 bound to D-PEPTIDE | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, D-Peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Noel, J.P, Zhang, Y. | | 登録日 | 2006-10-19 | | 公開日 | 2007-05-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural basis for high-affinity peptide inhibition of human Pin1.
Acs Chem.Biol., 2, 2007
|
|
2M09
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | 分子名称: | Splicing factor 1 | | 著者 | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | 登録日 | 2012-10-22 | | 公開日 | 2013-01-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
2M0G
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | 分子名称: | Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | 著者 | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | 登録日 | 2012-10-25 | | 公開日 | 2013-01-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
7T1F
 
 | |
3NTP
 
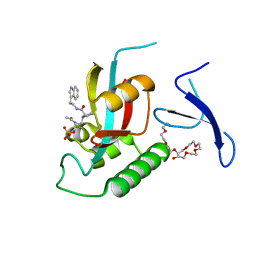 | | Human Pin1 complexed with reduced amide inhibitor | | 分子名称: | (2R)-2-(acetylamino)-3-[(2S)-2-{[2-(1H-indol-3-yl)ethyl]carbamoyl}pyrrolidin-1-yl]propyl dihydrogen phosphate, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Zhang, Y. | | 登録日 | 2010-07-05 | | 公開日 | 2012-01-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.762 Å) | | 主引用文献 | A reduced-amide inhibitor of Pin1 binds in a conformation resembling a twisted-amide transition state.
Biochemistry, 50, 2011
|
|
8H7G
 
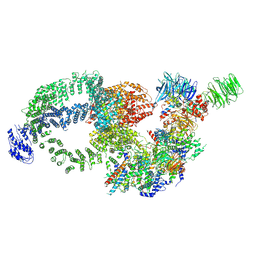 | |
3PWM
 
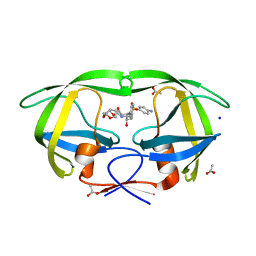 | | HIV-1 Protease Mutant L76V with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | 著者 | Zhang, Y, Weber, I.T. | | 登録日 | 2010-12-08 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
8HPB
 
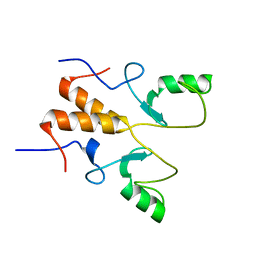 | |
8HQB
 
 | |
3PWR
 
 | | HIV-1 Protease Mutant L76V complexed with Saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Zhang, Y, Weber, I.T. | | 登録日 | 2010-12-08 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
3TH8
 
 | | Structure of E. coli undecaprenyl diphosphate synthase complexed with BPH-1063 | | 分子名称: | (2Z)-4-({3-[3-(hexyloxy)phenyl]propyl}amino)-2-hydroxy-4-oxobut-2-enoic acid, Undecaprenyl pyrophosphate synthase | | 著者 | Cao, R, Zhu, W, Zhang, Y, Oldfield, E. | | 登録日 | 2011-08-18 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.114 Å) | | 主引用文献 | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
3RY0
 
 | | Crystal structure of TomN, a 4-Oxalocrotonate Tautomerase homologue in Tomaymycin biosynthetic pathway | | 分子名称: | Putative tautomerase | | 著者 | Zhang, Y, Yan, W.P, Li, W.Z, Whitman, C.P. | | 登録日 | 2011-05-10 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Kinetic, Crystallographic, and Mechanistic Characterization of TomN: Elucidation of a Function for a 4-Oxalocrotonate Tautomerase Homologue in the Tomaymycin Biosynthetic Pathway.
Biochemistry, 50, 2011
|
|
4GA3
 
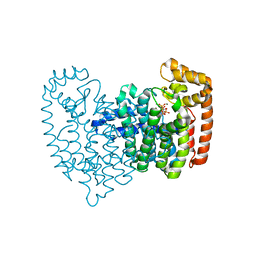 | | Crystal Structure of Human Farnesyl Diphosphate Synthase in Complex with BPH-1260 | | 分子名称: | 1-butyl-3-(2-hydroxy-2,2-diphosphonoethyl)-1H-imidazol-3-ium, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | 著者 | Liu, Y.-L, Zhang, Y, Oldfield, E. | | 登録日 | 2012-07-24 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Chemo-Immunotherapeutic Anti-Malarials Targeting Isoprenoid Biosynthesis.
ACS MED.CHEM.LETT., 4, 2013
|
|
4H6P
 
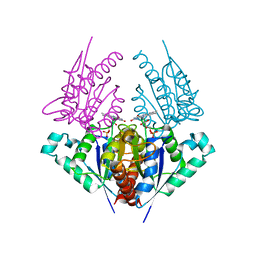 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a R101A substitution. | | 分子名称: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | 著者 | Zhang, Y, Robinson, H, Buchko, G.W. | | 登録日 | 2012-09-19 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.556 Å) | | 主引用文献 | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
7KHK
 
 | |
7KHJ
 
 | |
7KHG
 
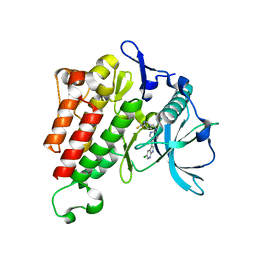 | | Crystal structure of KIT kinase domain with a small molecule inhibitor, PLX3397 | | 分子名称: | 5-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-{[6-(trifluoromethyl)pyridin-3-yl]methyl}pyridin-2-amine, Mast/stem cell growth factor receptor Kit | | 著者 | Zhang, Y. | | 登録日 | 2020-10-21 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Association of Combination of Conformation-Specific KIT Inhibitors With Clinical Benefit in Patients With Refractory Gastrointestinal Stromal Tumors: A Phase 1b/2a Nonrandomized Clinical Trial.
Jama Oncol, 7, 2021
|
|
4HS4
 
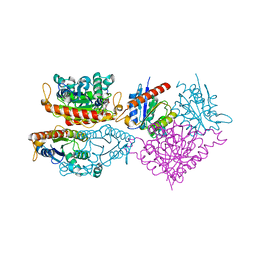 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a Y129N substitution. | | 分子名称: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | 著者 | Zhang, Y, Robinson, H, Buchko, G.W. | | 登録日 | 2012-10-29 | | 公開日 | 2012-12-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
5WAN
 
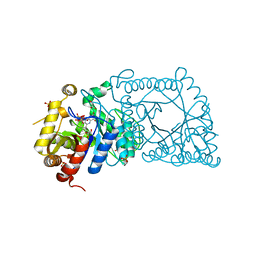 | | Crystal Structure of a flavoenzyme RutA in the pyrimidine catabolic pathway | | 分子名称: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Pyrimidine monooxygenase RutA, ... | | 著者 | Zhang, Y, Mukherjee, T, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | 登録日 | 2017-06-26 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | Catalysis of a flavoenzyme-mediated amide hydrolysis.
J. Am. Chem. Soc., 132, 2010
|
|
5X21
 
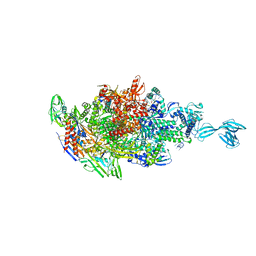 | | Crystal structure of Thermus thermophilus transcription initiation complex with GpA and pseudouridimycin (PUM) | | 分子名称: | (1S)-1,4-anhydro-5-[(N-carbamimidoylglycyl-N~2~-hydroxy-L-glutaminyl)amino]-5-deoxy-1-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-D-ribitol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Zhang, Y, Ebright, R. | | 登録日 | 2017-01-29 | | 公開日 | 2017-07-05 | | 最終更新日 | 2022-10-12 | | 実験手法 | X-RAY DIFFRACTION (3.323 Å) | | 主引用文献 | Antibacterial Nucleoside-Analog Inhibitor of Bacterial RNA Polymerase.
Cell, 169, 2017
|
|
5X22
 
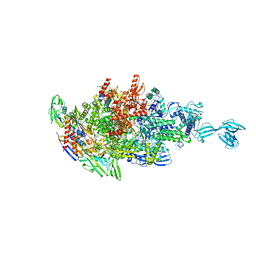 | |
5WVM
 
 | | Crystal structure of baeS cocrystallized with 2 mM indole | | 分子名称: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | 著者 | Wang, W, Zhang, Y, Rang, T, Xu, D. | | 登録日 | 2016-12-26 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
5WVN
 
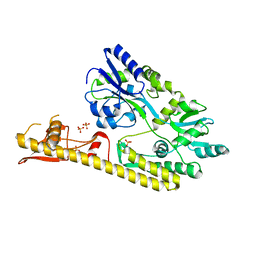 | | Crystal structure of MBS-BaeS fusion protein | | 分子名称: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | 著者 | Wang, W, Zhang, Y, Ran, T, Xu, D. | | 登録日 | 2016-12-26 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
3V7T
 
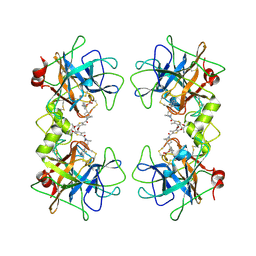 | |
6N33
 
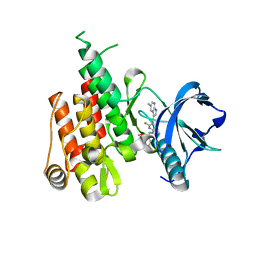 | | Crystal structure of fms kinase domain with a small molecular inhibitor, PLX5622 | | 分子名称: | 6-fluoro-N-[(5-fluoro-2-methoxypyridin-3-yl)methyl]-5-[(5-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]pyridin-2-amine, Macrophage colony-stimulating factor 1 receptor | | 著者 | Zhang, Y. | | 登録日 | 2018-11-14 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer's disease model.
Nat Commun, 10, 2019
|
|
