7VBP
 
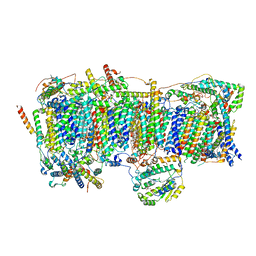 | | Membrane arm of deactive state CI from DQ-NADH dataset | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-09-01 | | 公開日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VBN
 
 | | Matrix arm of deactive state CI from DQ-NADH dataset | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-08-31 | | 公開日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VWL
 
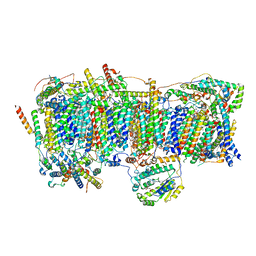 | | Membrane arm of deactive state CI from rotenone-NADH dataset | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-11-11 | | 公開日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VBZ
 
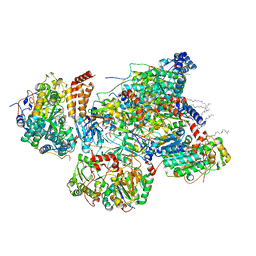 | | Matrix arm of active state CI from Rotenone-NADH dataset | | 分子名称: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-09-01 | | 公開日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VC0
 
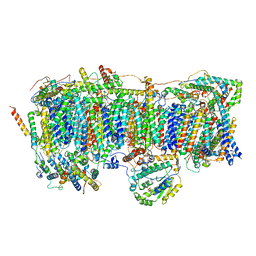 | | Membrane arm of active state CI from Rotenone-NADH dataset | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-09-01 | | 公開日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7YHQ
 
 | |
7YHP
 
 | |
7YHO
 
 | |
1HCF
 
 | | Crystal structure of TrkB-d5 bound to neurotrophin-4/5 | | 分子名称: | BDNF/NT-3 GROWTH FACTORS RECEPTOR, NEUROTROPHIN-4, SULFATE ION | | 著者 | Banfield, M.J, Naylor, R.L, Robertson, A.G.S, Allen, S.J, Dawbarn, D, Brady, R.L. | | 登録日 | 2001-05-03 | | 公開日 | 2001-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Specificity in Trk-Receptor:Neurotrophin Interaction: The Crystal Structure of Trkb-D5 in Complex with Neurotrophin-4/5
Structure, 9, 2001
|
|
4L91
 
 | | Crystal structure of Human Hsp90 with X29 | | 分子名称: | 4-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-6-chlorobenzene-1,3-diol, Heat shock protein HSP 90-alpha | | 著者 | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | 登録日 | 2013-06-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5Y4O
 
 | | Cryo-EM structure of MscS channel, YnaI | | 分子名称: | Low conductance mechanosensitive channel YnaI | | 著者 | Zhang, Y, Yu, J. | | 登録日 | 2017-08-04 | | 公開日 | 2019-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A binding-block ion selective mechanism revealed by a Na/K selective channel.
Protein Cell, 9, 2018
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | 分子名称: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | 著者 | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-08-12 | | 公開日 | 2013-09-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4L94
 
 | | Crystal structure of Human Hsp90 with S46 | | 分子名称: | (4-hydroxyphenyl)(4-methylpiperazin-1-yl)methanone, Heat shock protein HSP 90-alpha | | 著者 | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | 登録日 | 2013-06-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.649 Å) | | 主引用文献 | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5YC4
 
 | |
5YC3
 
 | |
4L8Z
 
 | | Crystal structure of Human Hsp90 with RL1 | | 分子名称: | Heat shock protein HSP 90-alpha, [5-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-2,4-dihydroxyphenyl](3,4-dihydroisoquinolin-2(1H)-yl)methanone | | 著者 | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | 登録日 | 2013-06-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.703 Å) | | 主引用文献 | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5Y20
 
 | |
7W1V
 
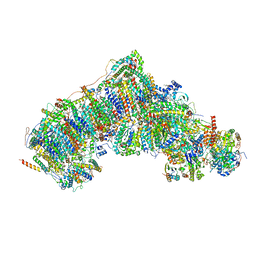 | | Active state CI from Rotenone-NADH dataset, Subclass 1 | | 分子名称: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-11-20 | | 公開日 | 2023-01-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W20
 
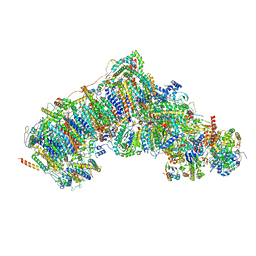 | | Active state CI from Rotenone-NADH dataset, Subclass 3 | | 分子名称: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-11-21 | | 公開日 | 2023-01-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VWJ
 
 | | Matrix arm of deactive state CI from rotenone-NADH dataset | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | 著者 | Gu, J.K, Yang, M.J. | | 登録日 | 2021-11-10 | | 公開日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XV9
 
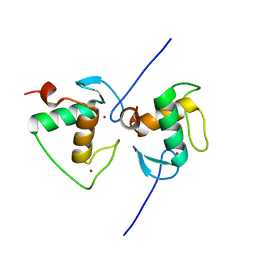 | | Crystal structure of the Human TR4 DNA-Binding Domain | | 分子名称: | Nuclear receptor subfamily 2 group C member 2, ZINC ION | | 著者 | Liu, Y, Chen, Z. | | 登録日 | 2022-05-21 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
7XV8
 
 | |
7XV6
 
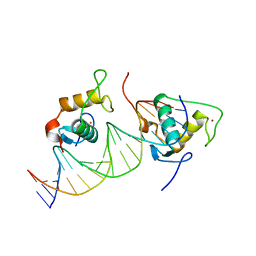 | |
7XVA
 
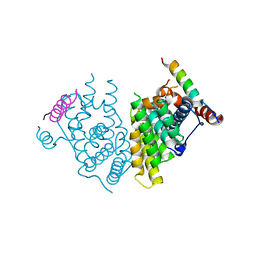 | |
7VJS
 
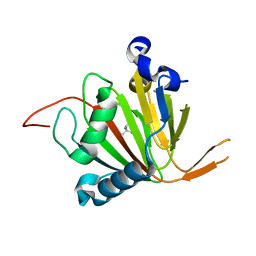 | | Human AlkB homolog ALKBH6 in complex with Tris and Ni | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, NICKEL (II) ION | | 著者 | Ma, L, Chen, Z. | | 登録日 | 2021-09-28 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
