6VOS
 
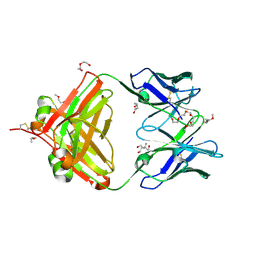 | | Crystal structure of macaque anti-HIV-1 antibody RM20J | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | 著者 | Yuan, M, Wilson, I.A. | | 登録日 | 2020-01-31 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
6Q9L
 
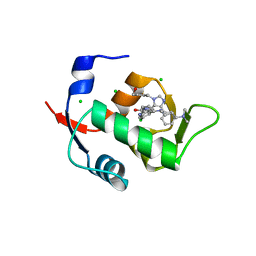 | |
6Q9U
 
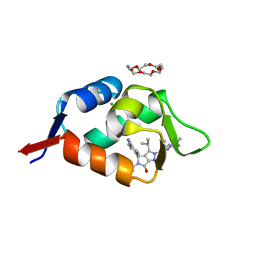 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 12 AT 2.4A; Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | 分子名称: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-[(4~{S})-5-(5-chloranyl-2-oxidanylidene-1~{H}-pyridin-3-yl)-2-[2-(dimethylamino)-4-methoxy-pyrimidin-5-yl]-6-oxidanylidene-3-propan-2-yl-4~{H}-pyrrolo[3,4-c]pyrazol-4-yl]benzenecarbonitrile, CHLORIDE ION, ... | | 著者 | Kallen, J. | | 登録日 | 2018-12-18 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9O
 
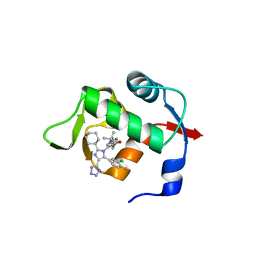 | |
6Q9Q
 
 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 13 AT 2.1A; Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | 分子名称: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 6-chloranyl-3-[3-[(1~{S})-1-(4-chlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-~{N}-[2-(4-cyclohexylpiperazin-1-yl)ethyl]-1~{H}-indole-2-carboxamide, GLYCEROL, ... | | 著者 | Kallen, J. | | 登録日 | 2018-12-18 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9W
 
 | | X-ray structure of compound 15 bound to HdmX: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | 分子名称: | (4~{S})-4-(4-chlorophenyl)-5-[(1~{S})-1-(3-chlorophenyl)ethyl]-2-(2,4-dimethoxypyrimidin-5-yl)-3-propan-2-yl-4~{H}-pyrrolo[3,4-d]imidazol-6-one, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Protein Mdm4, ... | | 著者 | Kallen, J. | | 登録日 | 2018-12-18 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q9H
 
 | |
6Q9Y
 
 | |
6Q9S
 
 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 14 AT 2.4A: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | 分子名称: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-[[6-chloranyl-3-[3-[(1~{S})-1-(2,4-dichlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-1~{H}-indol-2-yl]carbonylamino]-4-[4-(2-oxidanylidene-1,3-oxazinan-3-yl)piperidin-1-yl]benzoic acid, Protein Mdm4, ... | | 著者 | Kallen, J. | | 登録日 | 2018-12-18 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6Q96
 
 | |
5VAN
 
 | | Crystal Structure of Beta-Klotho | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Lee, S, Schlessinger, J. | | 登録日 | 2017-03-27 | | 公開日 | 2018-01-31 | | 最終更新日 | 2021-03-24 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
5VAK
 
 | | Crystal Structure of Beta-Klotho, Domain 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-klotho, ... | | 著者 | Lee, S, Schlessinger, J. | | 登録日 | 2017-03-27 | | 公開日 | 2018-01-31 | | 最終更新日 | 2021-03-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
5VAQ
 
 | |
1ZFP
 
 | |
7L0N
 
 | | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity | | 分子名称: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Snell, G, Czudnochowski, N, Dillen, J, Nix, J.C, Croll, T.I, Corti, D. | | 登録日 | 2020-12-11 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Cell, 184, 2021
|
|
5CT7
 
 | | BRAF in Complex with RAF265 | | 分子名称: | 1-methyl-5-({2-[5-(trifluoromethyl)-1H-imidazol-2-yl]pyridin-4-yl}oxy)-N-[4-(trifluoromethyl)phenyl]-1H-benzimidazol-2-amine, Serine/threonine-protein kinase B-raf | | 著者 | Appleton, B.A. | | 登録日 | 2015-07-23 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Discovery of RAF265: A Potent mut-B-RAF Inhibitor for the Treatment of Metastatic Melanoma.
Acs Med.Chem.Lett., 6, 2015
|
|
8OIZ
 
 | | Crystal structure of human CRBN-DDB1 in complex with Pomalidomide | | 分子名称: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Le Bihan, Y.-V, Cabry, M.P, van Montfort, R.L.M. | | 登録日 | 2023-03-23 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A Degron Blocking Strategy Towards Improved CRL4 CRBN Recruiting PROTAC Selectivity.
Chembiochem, 24, 2023
|
|
8OJH
 
 | | Crystal structure of human CRBN-DDB1 in complex with compound 4 | | 分子名称: | 1,2-ETHANEDIOL, 4-azanyl-2-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-7-methoxy-isoindole-1,3-dione, DNA damage-binding protein 1, ... | | 著者 | Cabry, M.P, Le Bihan, Y.-V, van Montfort, R.L.M. | | 登録日 | 2023-03-24 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | A Degron Blocking Strategy Towards Improved CRL4 CRBN Recruiting PROTAC Selectivity.
Chembiochem, 24, 2023
|
|
5V7Q
 
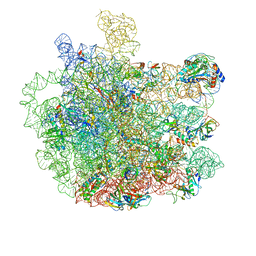 | | Cryo-EM structure of the large ribosomal subunit from Mycobacterium tuberculosis bound with a potent linezolid analog | | 分子名称: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Yang, K, Chang, J.-Y, Cui, Z, Zhang, J. | | 登録日 | 2017-03-20 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
7C0J
 
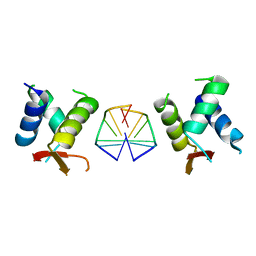 | |
7C0I
 
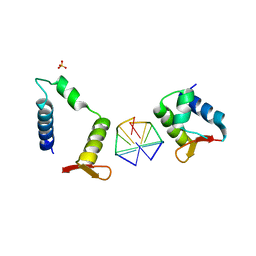 | | Crystal structure of chimeric mutant of E3L in complex with Z-DNA | | 分子名称: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-binding protein,Double-stranded RNA-specific adenosine deaminase, SULFATE ION | | 著者 | Choi, H.J, Park, C.H, Kim, J.S. | | 登録日 | 2020-05-01 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
7U0K
 
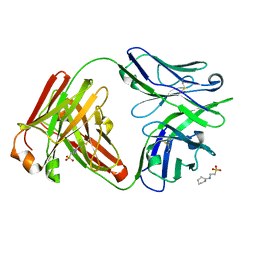 | | IOMA class antibody Fab ACS124 | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, IOMA Class antibody ACS124 Heavy chain, IOMA Class antibody ACS124 Light chain | | 著者 | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-02-18 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U04
 
 | | IOMA class antibody ACS101 | | 分子名称: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | 著者 | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-02-17 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U5G
 
 | | ACS122 Fab | | 分子名称: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | 著者 | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-03-02 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
6MAJ
 
 | | HBO1 is required for the maintenance of leukaemia stem cells | | 分子名称: | 4-fluoro-N'-[(3-hydroxyphenyl)sulfonyl]-5-methyl[1,1'-biphenyl]-3-carbohydrazide, BRD1 protein, GLYCEROL, ... | | 著者 | Ren, B, Peat, T.S, Monahan, B, Dawson, M, Street, I. | | 登録日 | 2018-08-27 | | 公開日 | 2019-12-25 | | 最終更新日 | 2020-01-22 | | 実験手法 | X-RAY DIFFRACTION (2.139 Å) | | 主引用文献 | HBO1 is required for the maintenance of leukaemia stem cells.
Nature, 577, 2020
|
|
