5KZC
 
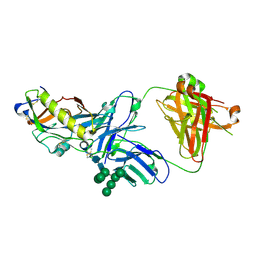 | | Crystal structure of an HIV-1 gp120 engineered outer domain with a Man9 glycan at position N276, in complex with broadly neutralizing antibody VRC01 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Engineered outer domain of gp120, VRC01 Fab heavy chain, ... | | 著者 | Julien, J.-P, Jardine, J.G, Diwanji, D, Schief, W.R, Wilson, I.A. | | 登録日 | 2016-07-24 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.
Plos Pathog., 12, 2016
|
|
3II5
 
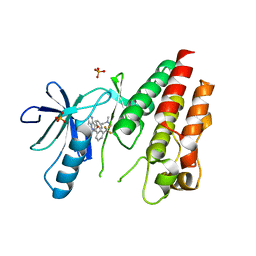 | | The Complex of wild-type B-RAF with Pyrazolo pyrimidine inhibitor | | 分子名称: | B-Raf proto-oncogene serine/threonine-protein kinase, N-[3-(3-{4-[(dimethylamino)methyl]phenyl}pyrazolo[1,5-a]pyrimidin-7-yl)phenyl]-3-(trifluoromethyl)benzamide, PHOSPHATE ION | | 著者 | Xu, W, Breger, D, Torres, N, Dutia, M, Powell, D, Ciszewski, G. | | 登録日 | 2009-07-31 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Non-hinge-binding pyrazolo[1,5-a]pyrimidines as potent B-Raf kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5APG
 
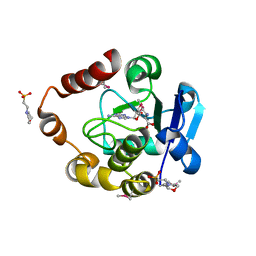 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from Vulcanisaeta distributa | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TSR3, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | 著者 | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | 登録日 | 2015-09-15 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
5LAW
 
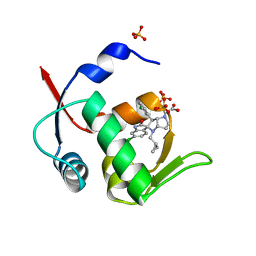 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND 14 | | 分子名称: | 2-[(3~{S},3'~{a}~{S},6'~{S},6'~{a}~{S})-6-chloranyl-6'-(3-chlorophenyl)-4'-(cyclopropylmethyl)-2-oxidanylidene-spiro[1~{H}-indole-3,5'-3,3~{a},6,6~{a}-tetrahydro-2~{H}-pyrrolo[3,2-b]pyrrole]-1'-yl]ethanoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | 著者 | Kessler, D, Gollner, A. | | 登録日 | 2016-06-15 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5LAZ
 
 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND BI-0252 | | 分子名称: | 4-[(2~{R},3~{a}~{S},5~{S},6~{S},6~{a}~{S})-6'-chloranyl-6-(3-chloranyl-2-fluoranyl-phenyl)-4-(cyclopropylmethyl)-2'-oxidanylidene-spiro[1,2,3,3~{a},6,6~{a}-hexahydropyrrolo[3,2-b]pyrrole-5,3'-1~{H}-indole]-2-yl]benzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION, ... | | 著者 | Kessler, D, Gollner, A. | | 登録日 | 2016-06-15 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
115D
 
 | | ORDERED WATER STRUCTURE IN AN A-DNA OCTAMER AT 1.7 ANGSTROMS RESOLUTION | | 分子名称: | DNA (5'-D(*GP*GP*(BRU)P*AP*(BRU)P*AP*CP*C)-3') | | 著者 | Kennard, O, Cruse, W.B.T, Nachman, J, Prange, T, Shakked, Z, Rabinovich, D. | | 登録日 | 1993-02-12 | | 公開日 | 1993-07-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Ordered water structure in an A-DNA octamer at 1.7 A resolution.
J.Biomol.Struct.Dyn., 3, 1986
|
|
1RVB
 
 | |
6HX9
 
 | | Putrescine transaminase from Pseudomonas putida | | 分子名称: | Aspartate aminotransferase family protein | | 著者 | Gahloth, D. | | 登録日 | 2018-10-16 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Characterization of a Putrescine Transaminase FromPseudomonas putidaand its Application to the Synthesis of Benzylamine Derivatives.
Front Bioeng Biotechnol, 6, 2018
|
|
5KWD
 
 | |
1GWL
 
 | | Carbohydrate binding module family29 complexed with mannohexaose | | 分子名称: | NON-CATALYTIC PROTEIN 1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | 著者 | Charnock, S.J, Nurizzo, D, Davies, G.J. | | 登録日 | 2002-03-19 | | 公開日 | 2003-03-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5LAV
 
 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) in complex with compound 6b | | 分子名称: | (3~{S},3'~{S},4'~{S})-4'-azanyl-6-chloranyl-3'-(3-chlorophenyl)-1'-(2,2-dimethylpropyl)spiro[1~{H}-indole-3,2'-pyrrolidine]-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | 著者 | Kessler, D, Gollner, A. | | 登録日 | 2016-06-15 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5LAY
 
 | | Discovery of New Natural-product-inspired Spiro-oxindole Compounds as Orally Active Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND 6g | | 分子名称: | (3~{S},3'~{S},4'~{S},5'~{S})-4'-azanyl-6-chloranyl-3'-(3-chloranyl-2-fluoranyl-phenyl)-1'-[(3-ethoxyphenyl)methyl]-5'-methyl-spiro[1~{H}-indole-3,2'-pyrrolidine]-2-one, E3 ubiquitin-protein ligase Mdm2, GLYCEROL, ... | | 著者 | Kessler, D, Gollner, A. | | 登録日 | 2016-06-15 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
6H9C
 
 | | Cryo-EM structure of archaeal extremophilic internal membrane-containing Haloarcula californiae icosahedral virus 1 (HCIV-1) at 3.74 Angstroms resolution. | | 分子名称: | (Half) GPS-II protein located underneath the two-tower capsomer sitting ON the icosahedral 2-fold axis., GPS-II protein located underneath the two-tower capsomer NOT sitting on the icosahedral 2-fold axis., GPS-III molecule located underneath the capsomer close to the icosahedral three-fold axis., ... | | 著者 | Abrescia, N.G, Santos-Perez, I, Charro, D. | | 登録日 | 2018-08-03 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Structural basis for assembly of vertical single beta-barrel viruses.
Nat Commun, 10, 2019
|
|
7W3H
 
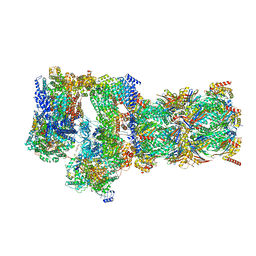 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.1_USP14 | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3B
 
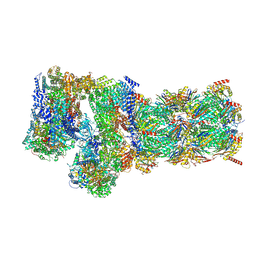 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED5_USP14 | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3G
 
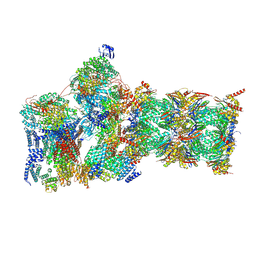 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.0_USP14 | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W39
 
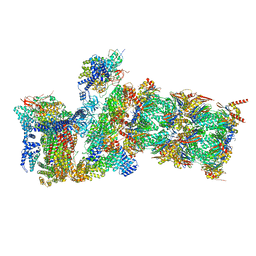 | | Structure of USP14-bound human 26S proteasome in state EA2.1_UBL | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W37
 
 | | Structure of USP14-bound human 26S proteasome in state EA1_UBL | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3C
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED0_USP14 | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3K
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD4_USP14 | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3J
 
 | | Structure of USP14-bound human 26S proteasome in substrate-inhibited state SC_USP14 | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3A
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED4_USP14 | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W3F
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED1_USP14 | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W38
 
 | | Structure of USP14-bound human 26S proteasome in state EA2.0_UBL | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
6I82
 
 | | Crystal structure of partially phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018412 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | 登録日 | 2018-11-19 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
