6UHX
 
 | | Crystal structure of YIR035C short chain dehydrogenases/reductase from Saccharomyces cerevisiae | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized oxidoreductase YIR035C | | 著者 | Stogios, P.J, Skarina, T, Chen, C, Kagan, O, Iakounine, A, Savchenko, A. | | 登録日 | 2019-09-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structure of YIR035C short chain dehydrogenases/reductase from Saccharomyces cerevisiae
To Be Published
|
|
5CXD
 
 | | 1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Holo-[acyl-carrier-protein] synthase, ... | | 著者 | Halavaty, A.S, Minasov, G, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-07-28 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | 1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group
To Be Published
|
|
5DT9
 
 | | Crystal structure of a putative D-Erythronate-4-Phosphate Dehydrogenase from Vibrio cholerae | | 分子名称: | CHLORIDE ION, Erythronate-4-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Stogios, P.J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-09-17 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.663 Å) | | 主引用文献 | Crystal structure of a putative D-Erythronate-4-Phosphate Dehydrogenase from Vibrio cholerae
To Be Published
|
|
7S2L
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 apoenzyme | | 分子名称: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
1KR4
 
 | | Structure Genomics, Protein TM1056, cutA | | 分子名称: | Protein TM1056, cutA | | 著者 | Savchenko, A, Zhang, R, Joachimiak, A, Edwards, A, Akarina, T, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-01-08 | | 公開日 | 2002-08-14 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-ray crystal structure of CutA from Thermotoga maritima at 1.4 A resolution.
Proteins, 54, 2004
|
|
1TD5
 
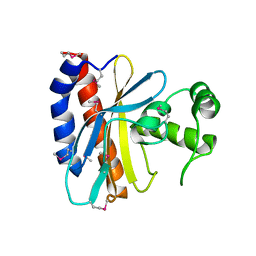 | | Crystal Structure of the Ligand Binding Domain of E. coli IclR. | | 分子名称: | Acetate operon repressor | | 著者 | Walker, J.R, Evdokimova, L, Zhang, R.-G, Bochkarev, A, Joachimiak, A, Arrowsmith, C, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-05-21 | | 公開日 | 2004-07-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Analyses of the Ligand Binding Sites of the IclR family of transcriptional regulators
To be Published
|
|
2PZ9
 
 | | Crystal structure of putative transcriptional regulator SCO4942 from Streptomyces coelicolor | | 分子名称: | Putative regulatory protein, SULFATE ION | | 著者 | Filippova, E.V, Chruszcz, M, Xu, X, Zheng, H, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-05-17 | | 公開日 | 2007-06-19 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | In situ proteolysis for protein crystallization and structure determination.
Nat.Methods, 4, 2007
|
|
6MN5
 
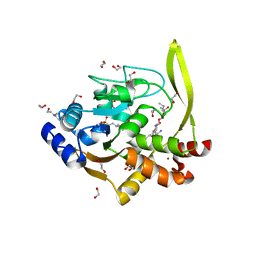 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, H154A mutant, in complex with gentamicin C1A | | 分子名称: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | 著者 | Stogios, P.J, Evdokimova, E, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-10-01 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MN4
 
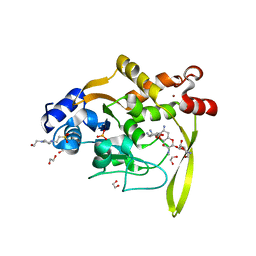 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, H154A mutant, in complex with apramycin | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, APRAMYCIN, ... | | 著者 | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-10-01 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MN3
 
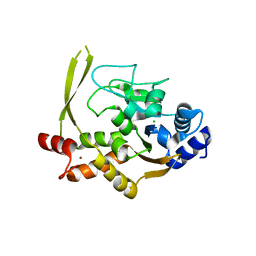 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, apoenzyme | | 分子名称: | Aminoglycoside N(3)-acetyltransferase, AAC(3)-IVa, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-10-01 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
1NI9
 
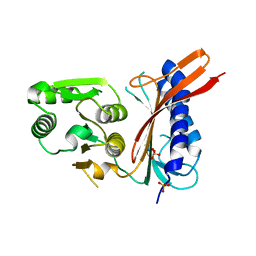 | | 2.0 A structure of glycerol metabolism protein from E. coli | | 分子名称: | Protein glpX, SULFATE ION | | 著者 | Sanishvili, R, Brunzelle, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-12-23 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
1QZ4
 
 | |
4RHA
 
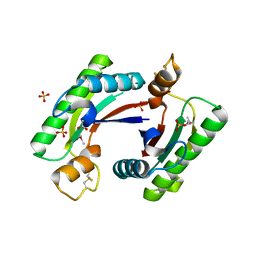 | | Structure of the C-terminal domain of outer-membrane protein OmpA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Outer membrane protein A, ... | | 著者 | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-10-01 | | 公開日 | 2014-10-29 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
2IS3
 
 | |
4JID
 
 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-free | | 分子名称: | CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein | | 著者 | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-03-05 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
2KKY
 
 | | Solution Structure of C-terminal domain of oxidized NleG2-3 (residue 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | 分子名称: | Uncharacterized protein ECs2156 | | 著者 | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | 登録日 | 2009-06-29 | | 公開日 | 2009-08-25 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
3N0M
 
 | | Crystal structure of BA2930 mutant (H183G) in complex with AcCoA | | 分子名称: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION | | 著者 | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-05-14 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3N0S
 
 | | Crystal structure of BA2930 mutant (H183A) in complex with AcCoA | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, ... | | 著者 | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-05-14 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
2KKX
 
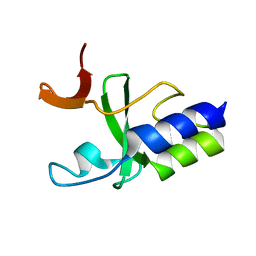 | | Solution Structure of C-terminal domain of reduced NleG2-3 (residues 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | 分子名称: | Uncharacterized protein ECs2156 | | 著者 | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-06-29 | | 公開日 | 2009-08-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
2LF3
 
 | | Solution NMR structure of HopPmaL_281_385 from Pseudomonas syringae pv. maculicola str. ES4326, Midwest Center for Structural Genomics target APC40104.5 and Northeast Structural Genomics Consortium target PsT2A | | 分子名称: | Effector protein hopAB3 | | 著者 | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | 登録日 | 2011-06-28 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
2LF6
 
 | | Solution NMR structure of HopABPph1448_220_320 from Pseudomonas syringae pv. phaseolicola str. 1448A, Midwest Center for Structural Genomics target APC40132.4 and Northeast Structural Genomics Consortium target PsT3A | | 分子名称: | Effector protein hopAB1 | | 著者 | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | 登録日 | 2011-06-28 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
6B5F
 
 | | Crystal structure of nicotinate mononucleotide-5,6-dimethylbenzimidazole phosphoribosyltransferase CobT from Yersinia enterocolitica | | 分子名称: | CHLORIDE ION, GLYCEROL, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, ... | | 著者 | Stogios, P.J, Skarina, T, McChesney, C, Grimshaw, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-09-29 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of nicotinate mononucleotide-5,6-dimethylbenzimidazole phosphoribosyltransferase CobT from Yersinia enterocolitica
To Be Published
|
|
3BRM
 
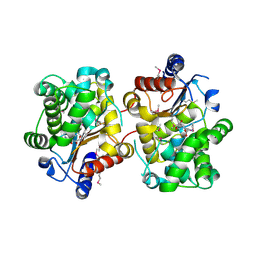 | | Crystal structure of the covalent complex between the Bacillus subtilis glutaminase YbgJ and 5-oxo-L-norleucine formed by reaction of the protein with 6-diazo-5-oxo-L-norleucine | | 分子名称: | 5-OXO-L-NORLEUCINE, Glutaminase 1 | | 著者 | Singer, A.U, Kim, Y, Dementieva, I, Vinokour, E, Joachimiak, A, Savchenko, A, Yakunin, A. | | 登録日 | 2007-12-21 | | 公開日 | 2008-05-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
1P9Q
 
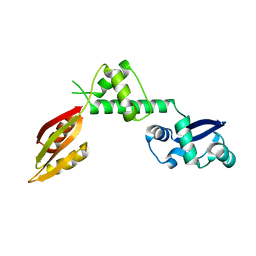 | | Structure of a hypothetical protein AF0491 from Archaeoglobus fulgidus | | 分子名称: | Hypothetical protein AF0491 | | 著者 | Savchenko, A, Evdokimova, E, Skarina, T, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A. | | 登録日 | 2003-05-12 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
5UXA
 
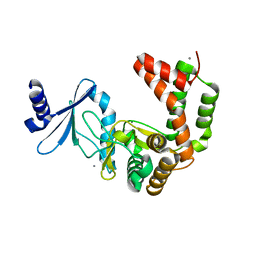 | | Crystal structure of macrolide 2'-phosphotransferase MphB from Escherichia coli | | 分子名称: | CALCIUM ION, Macrolide 2'-phosphotransferase II | | 著者 | Stogios, P.J, Evdokimova, E, Egorova, O, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-22 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
