7EP0
 
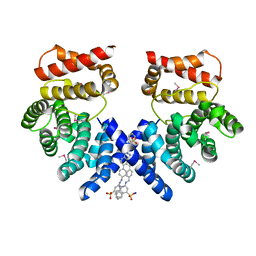 | | Crystal structure of ZYG11B bound to GSTE degron | | 分子名称: | Protein zyg-11 homolog B, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | 著者 | Yan, X, Li, Y. | | 登録日 | 2021-04-26 | | 公開日 | 2021-07-14 | | 最終更新日 | 2021-09-01 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP3
 
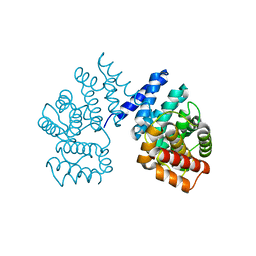 | |
1A6B
 
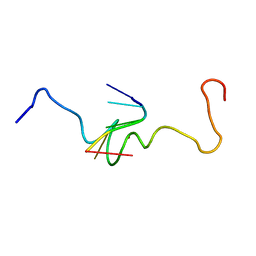 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE ZINC FINGER PROTEIN NCP10 OF MOLONEY MURINE LEUKEMIA VIRUS AND A SEQUENCE OF THE PSI-PACKAGING DOMAIN OF HIV-1, 20 STRUCTURES | | 分子名称: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), ZINC FINGER PROTEIN NCP10, ZINC ION | | 著者 | Schueler, W, Dong, C.-Z, Wecker, K, Roques, B.P. | | 登録日 | 1998-02-23 | | 公開日 | 1999-08-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes.
Biochemistry, 38, 1999
|
|
7E23
 
 | | SARS-CoV-2 spike in complex with the CA521 neutralizing antibody Fab (focused refinement on Fab-RBD) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CA521 Heavy Chain, CA521 Light Chain, ... | | 著者 | Liu, C, Song, D, Dou, C. | | 登録日 | 2021-02-04 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure and function analysis of a potent human neutralizing antibody CA521 FALA against SARS-CoV-2.
Commun Biol, 4, 2021
|
|
7Y8G
 
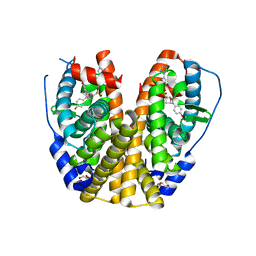 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with an Inhibitor 30a and GRIP Peptide | | 分子名称: | DI(HYDROXYETHYL)ETHER, Estrogen receptor, Grip peptide, ... | | 著者 | Min, J, Hu, H.B, Yang, Y, Dong, C.E, Zhou, H.B, Chen, C.-C, Guo, R.-T. | | 登録日 | 2022-06-23 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Structure-guided identification of novel dual-targeting estrogen receptor alpha degraders with aromatase inhibitory activity for the treatment of endocrine-resistant breast cancer.
Eur.J.Med.Chem., 253, 2023
|
|
7Y8F
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with an Inhibitor 30o and GRIP Peptide | | 分子名称: | DI(HYDROXYETHYL)ETHER, Estrogen receptor, Grip peptide, ... | | 著者 | Min, J, Hu, H.B, Yang, Y, Dong, C.E, Zhou, H.B, Chen, C.-C, Guo, R.-T. | | 登録日 | 2022-06-23 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structure-guided identification of novel dual-targeting estrogen receptor alpha degraders with aromatase inhibitory activity for the treatment of endocrine-resistant breast cancer.
Eur.J.Med.Chem., 253, 2023
|
|
5KCC
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with Oxabicyclic Heptene Sulfonamide (OBHS-N) | | 分子名称: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-phenyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2016-06-06 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.386 Å) | | 主引用文献 | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
8WWF
 
 | |
8WWE
 
 | |
8WWD
 
 | |
5CQR
 
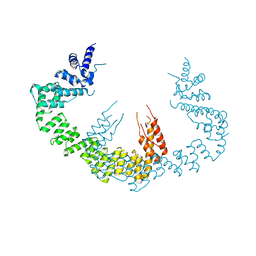 | | Dimerization of Elp1 is essential for Elongator complex assembly | | 分子名称: | Elongator complex protein 1 | | 著者 | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | 登録日 | 2015-07-22 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.015 Å) | | 主引用文献 | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CQS
 
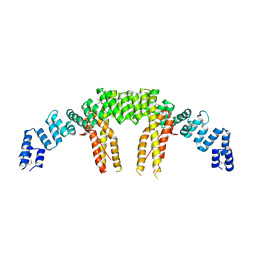 | | Dimerization of Elp1 is essential for Elongator complex assembly | | 分子名称: | Elongator complex protein 1 | | 著者 | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | 登録日 | 2015-07-22 | | 公開日 | 2015-08-19 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4O9B
 
 | | The Structure of CC1-IH in human STIM1. | | 分子名称: | CADMIUM ION, Stromal interaction molecule 1 | | 著者 | Cui, B, Yang, X, Li, S, Shen, Y. | | 登録日 | 2014-01-02 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.604 Å) | | 主引用文献 | The inhibitory helix controls the intramolecular conformational switching of the C-terminus of STIM1.
Plos One, 8, 2013
|
|
7WZC
 
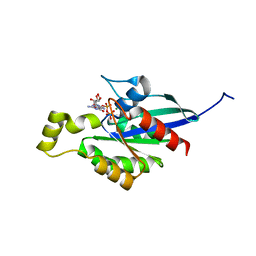 | |
7WZA
 
 | | An open conformation Form 1 of switch II for RhoA | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | 著者 | Jiang, H, Luo, C. | | 登録日 | 2022-02-17 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.50028777 Å) | | 主引用文献 | A RhoA structure with switch II flipped outward revealed the conformational dynamics of switch II region.
J.Struct.Biol., 215, 2023
|
|
7XCZ
 
 | |
7XDK
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7054 fab, ... | | 著者 | Liu, Z, Lui, S, Gao, Y. | | 登録日 | 2022-03-27 | | 公開日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDB
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7208 fab, ... | | 著者 | Liu, Z, Liu, S, Gao, Y.Z. | | 登録日 | 2022-03-26 | | 公開日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDA
 
 | |
7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | 著者 | Liu, Z, Liu, S, Yuanzhu, G. | | 登録日 | 2022-03-27 | | 公開日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XMW
 
 | | Crystal structure of anti-CRISPR protein AcrVIA2 | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AcrVIA2, SELENIUM ATOM, ... | | 著者 | Yan, X, Li, X, Song, G. | | 登録日 | 2022-04-27 | | 公開日 | 2023-05-31 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Structure of AcrVIA2 and its binding mechanism to CRISPR-Cas13a.
Biochem.Biophys.Res.Commun., 612, 2022
|
|
5EJM
 
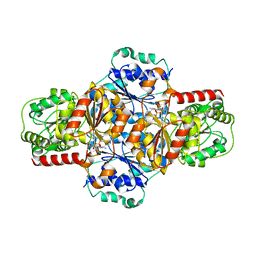 | | ThDP-Mn2+ complex of R413A variant of EcMenD soaked with 2-ketoglutarate for 35 min | | 分子名称: | (4~{R})-4-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3-ium-2-yl]-4-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | 著者 | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | 登録日 | 2015-11-02 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Two active site arginines are critical determinants of substrate binding and catalysis in MenD, a thiamine-dependent enzyme in menaquinone biosynthesis.
Biochem. J., 2018
|
|
7ONP
 
 | | Wild type carbonic anhydrase II with bound IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | 分子名称: | 4-[2-(9-chloranyl-2',3',4',5',6'-pentamethyl-4-nitro-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, ACETATE ION, Carbonic anhydrase 2, ... | | 著者 | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | 登録日 | 2021-05-25 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.408 Å) | | 主引用文献 | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7ONM
 
 | | Carbonic anhydrase II mutant (N67G-E69R-I91C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | 分子名称: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | 著者 | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | 登録日 | 2021-05-25 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.769 Å) | | 主引用文献 | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7ONQ
 
 | | Carbonic anhydrase II mutant (E69C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | 分子名称: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | 著者 | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | 登録日 | 2021-05-25 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
