7VZF
 
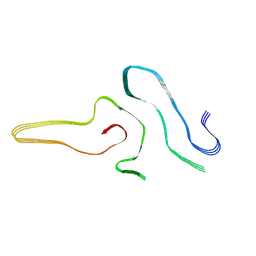 | | Cryo-EM structure of amyloid fibril formed by full-length human SOD1 | | 分子名称: | Superoxide dismutase [Cu-Zn] | | 著者 | Wang, L.Q, Ma, Y.Y, Yuan, H.Y, Zhao, K, Zhang, M.Y, Wang, Q, Huang, X, Xu, W.C, Chen, J, Li, D, Zhang, D.L, Zou, L.Y, Yin, P, Liu, C, Liang, Y. | | 登録日 | 2021-11-16 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Cryo-EM structure of an amyloid fibril formed by full-length human SOD1 reveals its conformational conversion.
Nat Commun, 13, 2022
|
|
6BSY
 
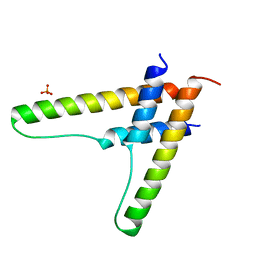 | | HIV-1 Rev assembly domain (residues 1-69) | | 分子名称: | PHOSPHATE ION, Protein Rev | | 著者 | Watts, N.R, Eren, E, Zhuang, X, Wang, Y.X, Steven, A.C, Wingfield, P.T. | | 登録日 | 2017-12-04 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A new HIV-1 Rev structure optimizes interaction with target RNA (RRE) for nuclear export.
J. Struct. Biol., 203, 2018
|
|
7XNE
 
 | | Crystal structure of CBP bromodomain liganded with Y08284 | | 分子名称: | CREB-binding protein, GLYCEROL, N-[3-(1-cyclopropylpyrazol-4-yl)-2-fluoranyl-5-[(1S)-1-oxidanylethyl]phenyl]-3-ethanoyl-7-methoxy-indolizine-1-carboxamide | | 著者 | Xiang, Q, Wang, C, Wu, T, Zhang, C, Hu, Q, Luo, G, Hu, J, Zhuang, X, Zou, L, Shen, H, Wu, X, Zhang, Y, Kong, X, Xu, Y. | | 登録日 | 2022-04-28 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
7WJS
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13157 | | 分子名称: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-07 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WKY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13153 | | 分子名称: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-12 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WMU
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13146 | | 分子名称: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, ~{N}-[4-[2,4-bis(fluoranyl)phenoxy]-3-[2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-5-methyl-4-oxidanylidene-furo[3,2-c]pyridin-7-yl]phenyl]ethanesulfonamide | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-17 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WNI
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13158 | | 分子名称: | 1,2-ETHANEDIOL, 7-[2-[2,4-bis(fluoranyl)phenoxy]-5-(2-oxidanylpropan-2-yl)phenyl]-2-[4-(2-hydroxyethyloxy)-3,5-dimethyl-phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-18 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WN5
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13142 | | 分子名称: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, POTASSIUM ION, ... | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-17 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WNA
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13120 | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Isoform 4 of Bromodomain-containing protein 2, ... | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-17 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WLN
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13153 | | 分子名称: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-13 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WMQ
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13157 | | 分子名称: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, FORMIC ACID, GLYCEROL, ... | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-16 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | 分子名称: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | 著者 | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | 登録日 | 2000-12-07 | | 公開日 | 2003-07-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
5GJR
 
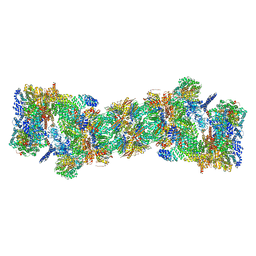 | | An atomic structure of the human 26S proteasome | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | 著者 | Huang, X.L, Luan, B, Wu, J.P, Shi, Y.G. | | 登録日 | 2016-07-01 | | 公開日 | 2016-09-07 | | 最終更新日 | 2019-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | An atomic structure of the human 26S proteasome.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5GJQ
 
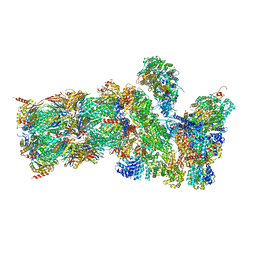 | | Structure of the human 26S proteasome bound to USP14-UbAl | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | 著者 | Huang, X.L, Luan, B, Wu, J.P, Shi, Y.G. | | 登録日 | 2016-07-01 | | 公開日 | 2016-08-17 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | An atomic structure of the human 26S proteasome.
Nat. Struct. Mol. Biol., 23, 2016
|
|
3U9N
 
 | |
7CFD
 
 | | Drosophila melanogaster Krimper eTud2-AubR15me2 complex | | 分子名称: | FI20010p1, Protein aubergine | | 著者 | Hu, H, Li, S. | | 登録日 | 2020-06-25 | | 公開日 | 2021-06-02 | | 最終更新日 | 2021-09-15 | | 実験手法 | X-RAY DIFFRACTION (2.704 Å) | | 主引用文献 | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFC
 
 | | Drosophila melanogaster Krimper eTud1-Ago3 complex | | 分子名称: | FI20010p1, Protein argonaute-3 | | 著者 | Hu, H, Li, S. | | 登録日 | 2020-06-25 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFB
 
 | | Drosophila melanogaster Krimper eTud1 apo structure | | 分子名称: | FI20010p1, SULFATE ION | | 著者 | Hu, H, Li, S. | | 登録日 | 2020-06-25 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7WFW
 
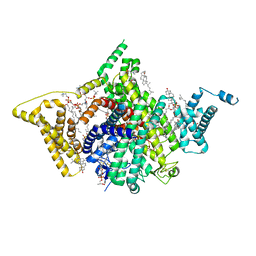 | | Apo human Nav1.8 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | 登録日 | 2021-12-27 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WEL
 
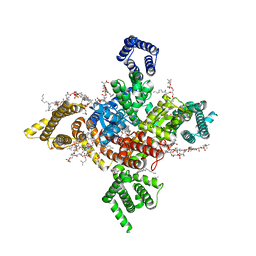 | | Human Nav1.8 with A-803467, class II | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | 登録日 | 2021-12-23 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WFR
 
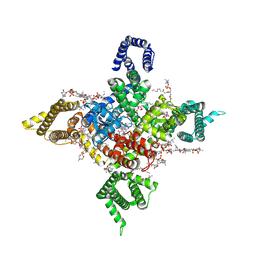 | | Human Nav1.8 with A-803467, class III | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | 登録日 | 2021-12-27 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WE4
 
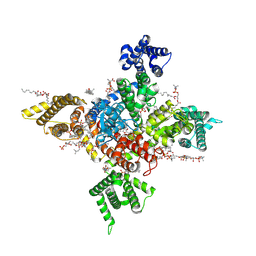 | | Human Nav1.8 with A-803467, class I | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | 登録日 | 2021-12-22 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5IO1
 
 | |
4AFX
 
 | | Crystal structure of the reactive loop cleaved ZPI in I2 space group | | 分子名称: | CALCIUM ION, PHOSPHATE ION, PROTEIN Z DEPENDENT PROTEASE INHIBITOR, ... | | 著者 | Zhou, A, Yan, Y, Wei, Z. | | 登録日 | 2012-01-23 | | 公開日 | 2013-01-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural Basis for Catalytic Activation of Protein Z-Dependent Protease Inhibitor (Zpi) by Protein Z.
Blood, 120, 2012
|
|
4AJU
 
 | | Crystal structure of the reactive loop cleaved ZPI in P41 space group | | 分子名称: | 1,2-ETHANEDIOL, PROTEIN Z-DEPENDENT PROTEASE INHIBITOR, SULFATE ION | | 著者 | Zhou, A, Yan, Y, Wei, Z. | | 登録日 | 2012-02-19 | | 公開日 | 2013-01-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural Basis for Catalytic Activation of Protein Z-Dependent Protease Inhibitor (Zpi) by Protein Z.
Blood, 120, 2012
|
|
