3E8E
 
 | | Crystal structures of the kinase domain of PKA in complex with ATP-competitive inhibitors | | 分子名称: | 4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, PKI inhibitor peptide, cAMP-dependent protein kinase catalytic subunit alpha | | 著者 | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | 登録日 | 2008-08-19 | | 公開日 | 2008-11-18 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3PKI
 
 | | Human SIRT6 crystal structure in complex with ADP ribose | | 分子名称: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | 著者 | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | 登録日 | 2010-11-11 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
3E88
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | 分子名称: | 4-[2-(4-amino-1,2,5-oxadiazol-3-yl)-6-{[(2R)-2-amino-3-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, Glycogen synthase kinase-3 beta peptide, RAC-beta serine/threonine-protein kinase | | 著者 | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | 登録日 | 2008-08-19 | | 公開日 | 2008-10-14 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3PKJ
 
 | | Human SIRT6 crystal structure in complex with 2'-N-Acetyl ADP ribose | | 分子名称: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | 著者 | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | 登録日 | 2010-11-11 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
3E87
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | 分子名称: | Glycogen synthase kinase-3 beta peptide, N-[(1S)-2-amino-1-phenylethyl]-5-(1H-pyrrolo[2,3-b]pyridin-4-yl)thiophene-2-carboxamide, RAC-beta serine/threonine-protein kinase | | 著者 | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | 登録日 | 2008-08-19 | | 公開日 | 2008-10-14 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1LMB
 
 | |
1LRP
 
 | |
8URU
 
 | | Spo11 core complex with hairpin DNA | | 分子名称: | Antiviral protein SKI8, Hairpin DNA, MAGNESIUM ION, ... | | 著者 | Yu, Y, Patel, D.J. | | 登録日 | 2023-10-26 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of the Spo11 core complex bound to DNA.
Nat.Struct.Mol.Biol., 2024
|
|
8URQ
 
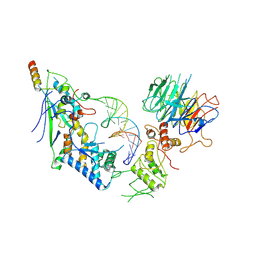 | | Spo11 core complex with gapped DNA | | 分子名称: | Antiviral protein SKI8, MAGNESIUM ION, Meiosis-specific protein SPO11, ... | | 著者 | Yu, Y, Patel, D.J. | | 登録日 | 2023-10-26 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures of the Spo11 core complex bound to DNA.
Nat.Struct.Mol.Biol., 2024
|
|
8WCP
 
 | |
6JQ5
 
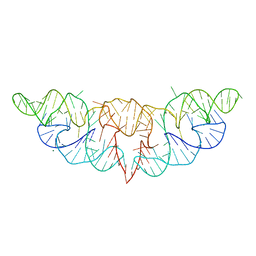 | | The structure of Hatchet Ribozyme | | 分子名称: | MAGNESIUM ION, RNA (82-MER) | | 著者 | Ren, A, Zheng, L. | | 登録日 | 2019-03-29 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.059 Å) | | 主引用文献 | Hatchet ribozyme structure and implications for cleavage mechanism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JG9
 
 | |
6JL7
 
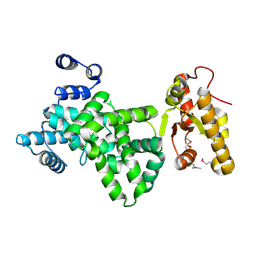 | |
6JQ6
 
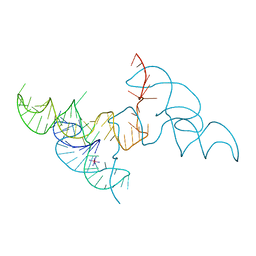 | |
6JG8
 
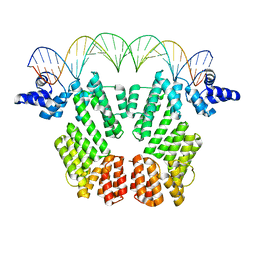 | |
7F2O
 
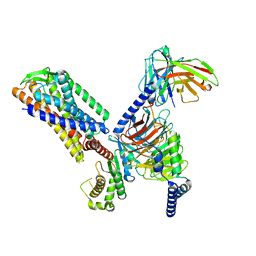 | |
6HBT
 
 | |
8VC8
 
 | |
6HBX
 
 | |
6HC5
 
 | |
7EIB
 
 | |
6JG5
 
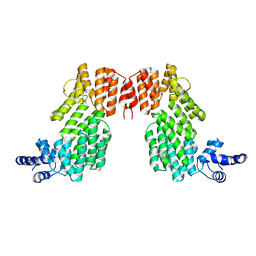 | | Crystal structure of AimR | | 分子名称: | AimR transcriptional regulator | | 著者 | Guan, Z.Y, Pei, K, Zou, T.T. | | 登録日 | 2019-02-13 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.221 Å) | | 主引用文献 | Structural insights into DNA recognition by AimR of the arbitrium communication system in the SPbeta phage.
Cell Discov, 5, 2019
|
|
3SQ4
 
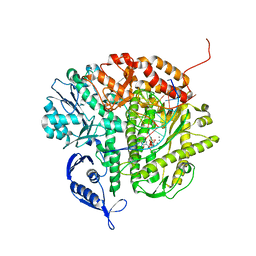 | | RB69 DNA Polymerase Ternary Complex with dTTP Opposite 2AP (GC rich sequence) | | 分子名称: | 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', CALCIUM ION, ... | | 著者 | Xia, S, Konigsberg, W.H, Wang, J. | | 登録日 | 2011-07-04 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SUQ
 
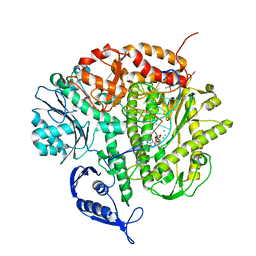 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dCTP Opposite 2AP (AT rich sequence) | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*G)-3', ... | | 著者 | Xia, S, Konigsberg, W.H, Wang, J. | | 登録日 | 2011-07-11 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
8T7I
 
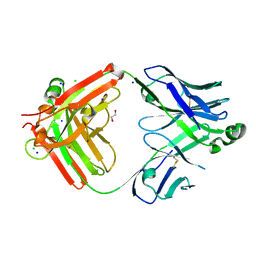 | | Structure of the S1CE variant of Fab F1 (FabS1CE-F1) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE variant of Fab F1 heavy chain, ... | | 著者 | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | 登録日 | 2023-06-20 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
