6IK9
 
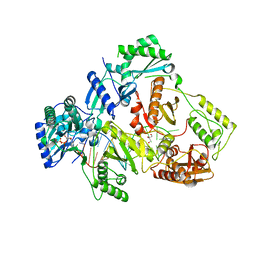 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:dGTP ternary complex | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2018-10-15 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.435 Å) | | 主引用文献 | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
8PEA
 
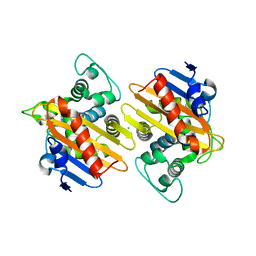 | |
7U55
 
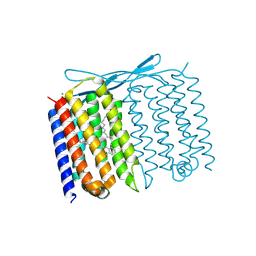 | | Crystal structure of Thermoplasmatales archaeon heliorhodopsin at pH 4.5 | | 分子名称: | CHLORIDE ION, DODECANE, Heliorhodopsin, ... | | 著者 | Besaw, J.E, De Guzman, P, Miller, R.J.D, Ernst, O.P. | | 登録日 | 2022-03-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Low pH structure of heliorhodopsin reveals chloride binding site and intramolecular signaling pathway.
Sci Rep, 12, 2022
|
|
6KDM
 
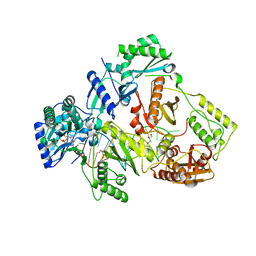 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:entecavir 5'-triphosphate ternary complex | | 分子名称: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2019-07-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDN
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dGTP ternary complex | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2019-07-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDJ
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:lamivudine 5'-triphosphate ternary complex | | 分子名称: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2019-07-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDK
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dCTP ternary complex | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2019-07-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDO
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y/M184V/F160M:DNA:lamivudine 5'-triphosphate ternary complex | | 分子名称: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2019-07-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.573 Å) | | 主引用文献 | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
3L6N
 
 | |
2ZIY
 
 | |
6G5W
 
 | | Crystal Structure of KDM4A with compound YP-03-038 | | 分子名称: | (4~{R})-5-methyl-4-phenyl-2-pyridin-2-yl-pyrazolidin-3-one, 1,2-ETHANEDIOL, CITRIC ACID, ... | | 著者 | Malecki, P.H, Carter, D.M, Gohlke, U, Specker, E, Nazare, M, Weiss, M.S, Heinemann, U. | | 登録日 | 2018-03-30 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Enhanced Properties of a Benzimidazole Benzylpyrazole Lysine Demethylase Inhibitor: Mechanism-of-Action, Binding Site Analysis, and Activity in Cellular Models of Prostate Cancer.
J.Med.Chem., 64, 2021
|
|
6G5X
 
 | | Crystal Structure of KDM4A with compound YP-02-145 | | 分子名称: | 1,2-ETHANEDIOL, 2-(3-methyl-5-oxidanylidene-4-phenyl-4~{H}-pyrazol-1-yl)-3~{H}-benzimidazole-5-carboxylic acid, CITRIC ACID, ... | | 著者 | Malecki, P.H, Carter, D.M, Gohlke, U, Specker, E, Nazare, M, Weiss, M.S, Heinemann, U. | | 登録日 | 2018-03-30 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Enhanced Properties of a Benzimidazole Benzylpyrazole Lysine Demethylase Inhibitor: Mechanism-of-Action, Binding Site Analysis, and Activity in Cellular Models of Prostate Cancer.
J.Med.Chem., 64, 2021
|
|
7K41
 
 | | Bacterial O-GlcNAcase (OGA) with compound | | 分子名称: | 1,2-ETHANEDIOL, 4-(4-methylpiperidin-1-yl)-N-(2-phenylethyl)pyrimidin-2-amine, ACETATE ION, ... | | 著者 | Lane, W, Tjhen, R, Snell, G, Sang, B. | | 登録日 | 2020-09-14 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of a Novel and Brain-Penetrant O -GlcNAcase Inhibitor via Virtual Screening, Structure-Based Analysis, and Rational Lead Optimization.
J.Med.Chem., 64, 2021
|
|
2E5S
 
 | | Solution structure of the zf-CCCHx2 domain of muscleblind-like 2, isoform 1 [Homo sapiens] | | 分子名称: | OTTHUMP00000018578, ZINC ION | | 著者 | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-12-22 | | 公開日 | 2007-06-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
8OXI
 
 | |
8OXH
 
 | |
8OXL
 
 | |
8PHY
 
 | |
8OXK
 
 | |
8OXJ
 
 | |
8PEC
 
 | | OXA-48_Q5-CAZ. Epistasis Arises from Shifting the Rate-Limiting Step during Enzyme Evolution | | 分子名称: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase, CHLORIDE ION | | 著者 | Leiros, H.-K.S, Frohlich, C. | | 登録日 | 2023-06-13 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Epistasis arises from shifting the rate-limiting step during enzyme evolution of a beta-lactamase.
Nat Catal, 7, 2024
|
|
8PEB
 
 | |
2DK4
 
 | | Solution structure of Splicing Factor Motif in Pre-mRNA splicing factor 18 (hPRP18) | | 分子名称: | Pre-mRNA-splicing factor 18 | | 著者 | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-04-06 | | 公開日 | 2006-10-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the splicing factor motif of the human Prp18 protein.
Proteins, 80, 2012
|
|
3AY4
 
 | | Crystal structure of nonfucosylated Fc complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, Low affinity immunoglobulin gamma Fc region receptor III-A, ... | | 著者 | Mizushima, T, Takemoto, E, Yagi, H, Shibata-Koyama, M, Isoda, Y, Iida, S, Satoh, M, Kato, K. | | 登録日 | 2011-04-28 | | 公開日 | 2011-08-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans
Genes Cells, 16, 2011
|
|
2LWI
 
 | | Solution structure of H-RasT35S mutant protein in complex with Kobe2601 | | 分子名称: | 2-(2,4-dinitrophenyl)-N-(4-fluorophenyl)hydrazinecarbothioamide, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Araki, M, Tamura, A, Shima, F, Kataoka, T. | | 登録日 | 2012-08-01 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
