8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | 分子名称: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | 登録日 | 2023-02-12 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | 分子名称: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | 登録日 | 2023-02-12 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | 分子名称: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | 登録日 | 2023-02-12 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | 分子名称: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | 登録日 | 2023-02-12 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
6PZV
 
 | |
3KPH
 
 | |
7RW2
 
 | |
4TKQ
 
 | | Native-SAD phasing for YetJ from Bacillus Subtilis | | 分子名称: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein YetJ | | 著者 | Liu, Q, Chang, Y, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2014-05-27 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8025 Å) | | 主引用文献 | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5WUC
 
 | | Structural basis for conductance through TRIC cation channels | | 分子名称: | SODIUM ION, Uncharacterized protein | | 著者 | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2016-12-17 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUD
 
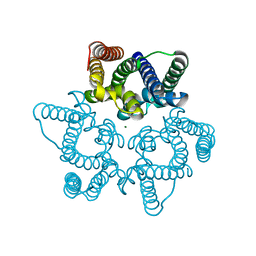 | | Structural basis for conductance through TRIC cation channels | | 分子名称: | MAGNESIUM ION, Uncharacterized protein | | 著者 | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2016-12-17 | | 公開日 | 2017-06-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUE
 
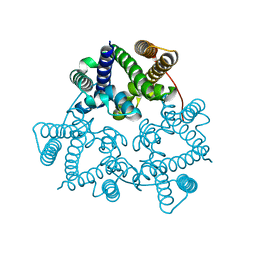 | | Structural basis for conductance through TRIC cation channels | | 分子名称: | SULFATE ION, Uncharacterized protein | | 著者 | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2016-12-17 | | 公開日 | 2017-06-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
6M17
 
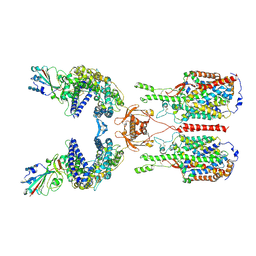 | | The 2019-nCoV RBD/ACE2-B0AT1 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Guo, Y.Y, Zhou, Q. | | 登録日 | 2020-02-24 | | 公開日 | 2020-03-11 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
7PBI
 
 | |
7PBG
 
 | |
8HWG
 
 | | D5 ATPrS-ADP-ssDNA form | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | 著者 | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | 登録日 | 2022-12-29 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWH
 
 | | Cryo-EM Structure of D5 Apo-ssDNA form | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), Primase D5 | | 著者 | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | 登録日 | 2022-12-29 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWD
 
 | | Cryo-EM Structure of D5 ADP form | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Primase D5 | | 著者 | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | 登録日 | 2022-12-29 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWC
 
 | | Cryo-EM Structure of D5 Apo | | 分子名称: | Primase D5 | | 著者 | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | 登録日 | 2022-12-29 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWB
 
 | | D5 ATP-ADP-Apo-ssDNA IS2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | 著者 | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | 登録日 | 2022-12-29 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWA
 
 | | D5 ATP-ADP-Apo-ssDNA IS1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | 著者 | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | 登録日 | 2022-12-29 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWF
 
 | | Cryo-EM Structure of D5 ADP-ssDNA form | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | 著者 | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | 登録日 | 2022-12-29 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HWE
 
 | | Cryo-EM Structure of D5 ATP-ADP form | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | 登録日 | 2022-12-29 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5KF4
 
 | |
7MHS
 
 | | Structure of p97 (subunits A to E) with substrate engaged | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Xu, Y, Han, H, Cooney, I, Hill, C.P, Shen, P.S. | | 登録日 | 2021-04-15 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Active conformation of the p97-p47 unfoldase complex.
Nat Commun, 13, 2022
|
|
6UBI
 
 | |
