2JOO
 
 | | The NMR Solution Structure of Recombinant RGD-hirudin | | Descriptor: | Hirudin variant-1 | | Authors: | Song, X, Mo, W, Liu, X, Yan, X, Song, H, Dai, L. | | Deposit date: | 2007-03-14 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of recombinant RGD-hirudin
Biochem.Biophys.Res.Commun., 360, 2007
|
|
8D1T
 
 | | Crystal structure of human USP30 in complex with a covalent inhibitor 552 and a Fab | | Descriptor: | (1R,2R,4S,7E)-7-[amino(sulfanyl)methylidene]-2-{[(1P)-3-chloro-3'-(1-cyanocyclopropyl)[1,1'-biphenyl]-4-carbonyl]amino}-7-azabicyclo[2.2.1]heptan-7-ium, 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 30, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | TBD
To Be Published
|
|
8D0A
 
 | | Crystal structure of human USP30 in complex with a covalent inhibitor 829 and a Fab | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 30, ZINC ION, mouse anti-huUSP30 Fab heavy chain, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | TBD
To Be Published
|
|
5UN1
 
 | | Crystal structure of GluN1/GluN2B delta-ATD NMDA receptor | | Descriptor: | (5S,10R)-5-methyl-10,11-dihydro-5H-5,10-epiminodibenzo[a,d][7]annulene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, X, Gouaux, E. | | Deposit date: | 2017-01-30 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanism of NMDA receptor channel block by MK-801 and memantine.
Nature, 556, 2018
|
|
8SHB
 
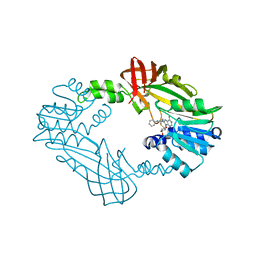 | | Crystal Structure of PRMT3 with Compound YD1-208 | | Descriptor: | 5'-S-[3-(N'-phenylcarbamimidamido)propyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of PRMT3 with Compound YD1-208
To be published
|
|
8SP6
 
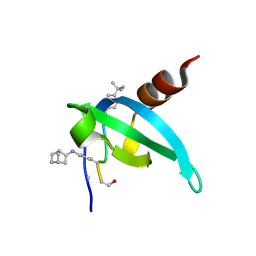 | | COMPLEX STRUCTURE OF CDYL2 WITH AN ANTAGONIST | | Descriptor: | (1s,4s)-bicyclo[2.2.1]heptane, (5R0)FAL(MLZ)(5R5), Chromodomain Y-like protein 2, ... | | Authors: | Song, X, Beldar, S, Dong, A, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | COMPLEX STRUCTURE OF CDYL2 WITH AN ANTAGONIST
To be published
|
|
8SIG
 
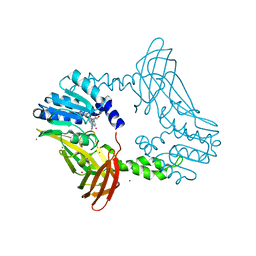 | | Crystal Structure of PRMT4 with Compound YD1-288 | | Descriptor: | 5'-{(3-aminopropyl)[2-(benzylcarbamamido)ethyl]amino}-5'-deoxyadenosine, Histone-arginine methyltransferase CARM1, SODIUM ION, ... | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of PRMT4 with Compound YD1-288
To be published
|
|
8SIH
 
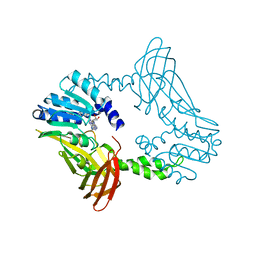 | | Crystal Structure of PRMT4 with Compound YD1-289 | | Descriptor: | 5'-{[2-(benzylcarbamamido)ethyl][3-(N'-cyclopentylcarbamimidamido)propyl]amino}-5'-deoxyadenosine, CALCIUM ION, Histone-arginine methyltransferase CARM1 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of PRMT4 with Compound YD1-289
To be published
|
|
8SHR
 
 | | Crystal Structure of PRMT3 with Compound YD1-214 | | Descriptor: | 5'-S-[2-(phenylcarbamamido)ethyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of PRMT3 with Compound YD1-214
To be published
|
|
8SIO
 
 | | Crystal structure of PRMT3 with YD1-66 | | Descriptor: | 5'-S-{3-[N'-(4'-chloro[1,1'-biphenyl]-3-yl)carbamimidamido]propyl}-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PRMT3 with YD1-66
To be published
|
|
8SII
 
 | | Crystal Structure of CBX7 with compound UNC4976 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Chromobox protein homolog 7, ... | | Authors: | Song, X, Dong, A, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-16 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structure of CBX7 with compound UNC4976
To be published
|
|
7SLZ
 
 | | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH BPF023596 | | Descriptor: | Glucose-induced degradation protein 4 homolog, N-[(1s,4s)-4-(1H-benzimidazol-2-yl)cyclohexyl]-N~2~-[(1H-indol-2-yl)methyl]glycinamide | | Authors: | Song, X, Dong, A, Calabrese, M, Wang, F, Owen, D, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH BPF023596
To Be Published
|
|
4F7G
 
 | |
8IA1
 
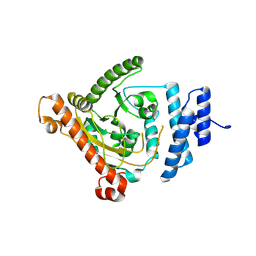 | |
6JN2
 
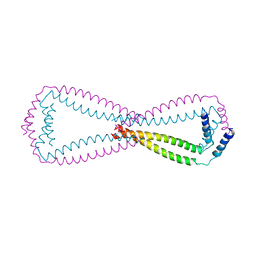 | | Crystal structure of the coiled-coil domains of human DOT1L in complex with AF10 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, Protein AF-10 | | Authors: | Song, X, Wang, M, Yang, N, Xu, R.M. | | Deposit date: | 2019-03-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A higher-order configuration of the heterodimeric DOT1L-AF10 coiled-coil domains potentiates their leukemogenenic activity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JC3
 
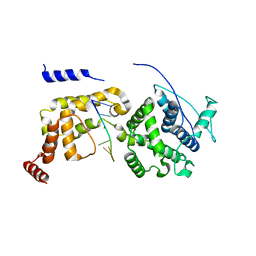 | | The Cryo-EM structure of nucleoprotein-RNA complex of Newcastle disease virus | | Descriptor: | Nucleocapsid, polyU | | Authors: | Song, X, Shan, H, Zhu, Y, Ding, W, Ouyang, S, Shen, Q.T, Liu, Z.J. | | Deposit date: | 2019-01-28 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Self-capping of nucleoprotein filaments protects the Newcastle disease virus genome.
Elife, 8, 2019
|
|
7CYC
 
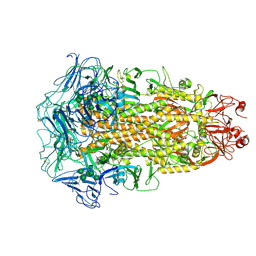 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
7CYD
 
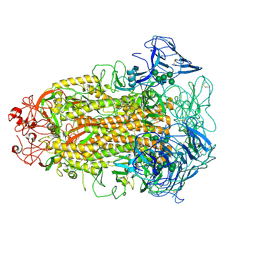 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
6M1I
 
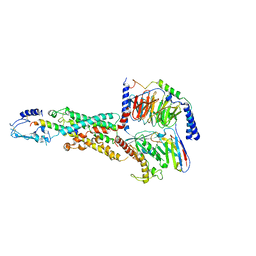 | | CryoEM structure of human PAC1 receptor in complex with PACAP38 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
4OUL
 
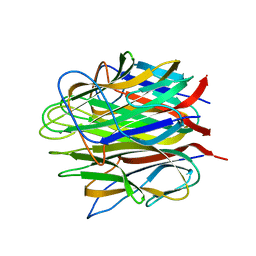 | | Crystal structure of human Caprin-2 C1q domain | | Descriptor: | CALCIUM ION, Caprin-2, GLYCEROL | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-17 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
4OUS
 
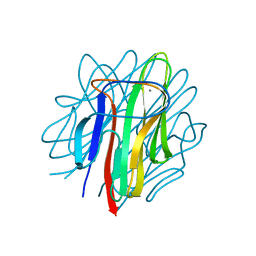 | | Crystal structure of zebrafish Caprin-2 C1q domain | | Descriptor: | CALCIUM ION, Caprin-2 | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
4OUM
 
 | | Crystal structure of human Caprin-2 C1q domain | | Descriptor: | CITRATE ANION, Caprin-2, ISOPROPYL ALCOHOL, ... | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
8G2F
 
 | | Crystal Structure of PRMT3 with Compound II710 | | Descriptor: | 5'-S-[3-(N'-benzylcarbamimidamido)propyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
8G2G
 
 | | Crystal structure of PRMT3 with compound YD1113 | | Descriptor: | 5'-S-[2-(benzylcarbamamido)ethyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3, SULFATE ION | | Authors: | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | Deposit date: | 2023-02-03 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A unique binding pocket induced by a noncanonical SAH mimic to develop potent and selective PRMT inhibitors.
Acta Pharm Sin B, 13, 2023
|
|
