4V0I
 
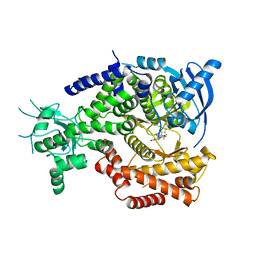 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | Deposit date: | 2014-09-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
6YHS
 
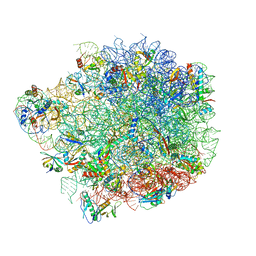 | | Acinetobacter baumannii ribosome-amikacin complex - 50S subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-03-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
6YPU
 
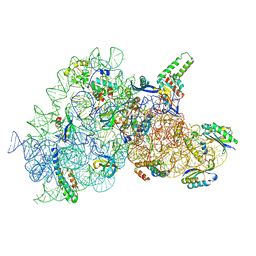 | | Acinetobacter baumannii ribosome-amikacin complex - 30S subunit body | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, 16S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
6YS5
 
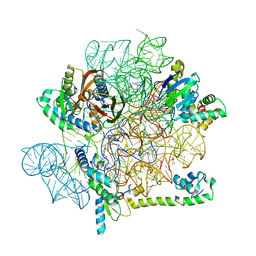 | | Acinetobacter baumannii ribosome-amikacin complex - 30S subunit head | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
6YTF
 
 | | Acinetobacter baumannii ribosome-tigecycline complex - 30S subunit head | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-24 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
6YT9
 
 | | Acinetobacter baumannii ribosome-tigecycline complex - 30S subunit body | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-24 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
7QEP
 
 | | Cryo-EM structure of the ribosome from Encephalitozoon cuniculi | | Descriptor: | 18S ribosomal RNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Nicholson, D, Ranson, N.A, Melnikov, S.V. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Adaptation to genome decay in the structure of the smallest eukaryotic ribosome
Nat Commun, 13, 2022
|
|
6YSI
 
 | | Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
1T2X
 
 | | Glactose oxidase C383S mutant identified by directed evolution | | Descriptor: | ACETATE ION, COPPER (II) ION, Galactose Oxidase, ... | | Authors: | Wilkinson, D, Akumanyi, N, Hurtado-Guerrero, R, Dawkes, H, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies of a series of mutants of galactose oxidase identified by directed evolution.
Protein Eng.Des.Sel., 17, 2004
|
|
4CBQ
 
 | | Crystal structure of the thioredoxin reductase from Entamoeba histolytica with auranofin Au(I) bound to Cys286 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, ... | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-16 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4CW9
 
 | | Entamoeba histolytica thiredoxin C34S mutant | | Descriptor: | THIOREDOXIN | | Authors: | Parsonage, D, Kells, P.M, Vieira, D.F, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2014-04-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4CCR
 
 | | Crystal structure of the thioredoxin reductase apoenzyme from Entamoeba histolytica in the absence of the NADP cofactor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, THIOREDOXIN REDUCTASE | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-25 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
1GYK
 
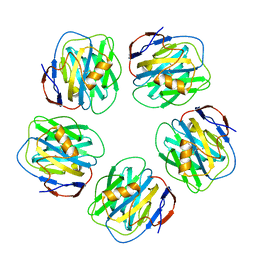 | | Serum Amyloid P Component co-crystallised with MOBDG at neutral pH | | Descriptor: | CALCIUM ION, METHYL 4,6-O-[(1R)-1-CARBOXYETHYLIDENE]-BETA-D-GALACTOPYRANOSIDE, SERUM AMYLOID P-COMPONENT | | Authors: | Thompson, D, Pepys, M.B, Tickle, I, Wood, S.P. | | Deposit date: | 2002-04-25 | | Release date: | 2003-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structures of Crystalline Complexes of Human Serum Amyloid P Component with its Carbohydrate Ligand, the Cyclic Pyruvate Acetal of Galactose
J.Mol.Biol., 320, 2002
|
|
4CCQ
 
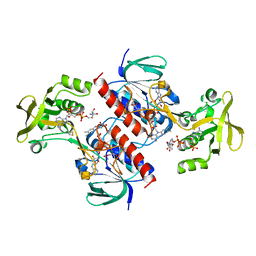 | | Crystal structure of the thioredoxin reductase from Entamoeba histolytica with NADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
1YF0
 
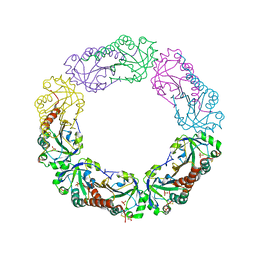 | | Structural and biochemical analysis of the link between enzymatic activity and oligomerization in AhpC, a bacterial peroxiredoxin. | | Descriptor: | Alkyl hydroperoxide reductase subunit C, SULFATE ION | | Authors: | Parsonage, D, Youngblood, D.S, Sarma, G.N, Wood, Z.A, Karplus, P.A, Poole, L.B. | | Deposit date: | 2004-12-29 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis of the Link between Enzymatic Activity and Oligomeric State in AhpC, a Bacterial Peroxiredoxin.
Biochemistry, 44, 2005
|
|
2JD4
 
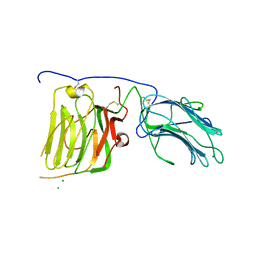 | | Mouse laminin alpha1 chain, domains LG4-5 | | Descriptor: | CHLORIDE ION, LAMININ SUBUNIT ALPHA-1, MAGNESIUM ION | | Authors: | Harrison, D, Hussain, S.A, Combs, A.C, Ervasti, J.M, Yurchenco, P.D, Hohenester, E. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Cell Surface Anchorage Sites of Laminin {Alpha}1Lg4-5.
J.Biol.Chem., 282, 2007
|
|
1YF1
 
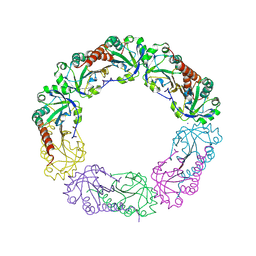 | | Structural and biochemical analysis of the link between enzymatic activity and oligomerization in AhpC, a bacterial peroxiredoxin. | | Descriptor: | Alkyl hydroperoxide reductase subunit C, SODIUM ION | | Authors: | Parsonage, D, Youngblood, D.S, Sarma, G.N, Wood, Z.A, Karplus, P.A, Poole, L.B. | | Deposit date: | 2004-12-29 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of the Link between Enzymatic Activity and Oligomeric State in AhpC, a Bacterial Peroxiredoxin.
Biochemistry, 44, 2005
|
|
1YEX
 
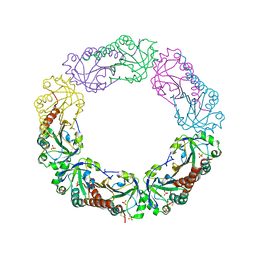 | | Structural and biochemical analysis of the link between enzymatic activity and oligomerization in AhpC, a bacterial peroxiredoxin. | | Descriptor: | Alkyl hydroperoxide reductase subunit C, SULFATE ION | | Authors: | Parsonage, D, Youngblood, D.S, Sarma, G.N, Wood, Z.A, Karplus, P.A, Poole, L.B. | | Deposit date: | 2004-12-28 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Analysis of the Link between Enzymatic Activity and Oligomeric State in AhpC, a Bacterial Peroxiredoxin.
Biochemistry, 44, 2005
|
|
1YEP
 
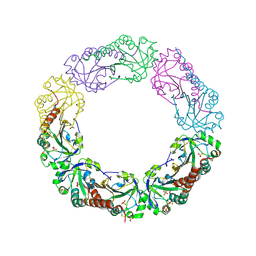 | | Structural and biochemical analysis of the link between enzymatic activity and olgomerization in AhpC, a bacterial peroxiredoxin. | | Descriptor: | Alkyl hydroperoxide reductase subunit C, SULFATE ION | | Authors: | Parsonage, D, Youngblood, D.S, Sarma, G.N, Wood, Z.A, Karplus, P.A, Poole, L.B. | | Deposit date: | 2004-12-28 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis of the Link between Enzymatic Activity and Oligomeric State in AhpC, a Bacterial Peroxiredoxin.
Biochemistry, 44, 2005
|
|
1B09
 
 | |
7P48
 
 | | Staphylococcus aureus ribosome in complex with Sal(B) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nicholson, D, Ranson, N.A, O'Neill, A.J. | | Deposit date: | 2021-07-09 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Sal-type ABC-F proteins: intrinsic and common mediators of pleuromutilin resistance by target protection in staphylococci.
Nucleic Acids Res., 50, 2022
|
|
7E23
 
 | | SARS-CoV-2 spike in complex with the CA521 neutralizing antibody Fab (focused refinement on Fab-RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CA521 Heavy Chain, CA521 Light Chain, ... | | Authors: | Liu, C, Song, D, Dou, C. | | Deposit date: | 2021-02-04 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and function analysis of a potent human neutralizing antibody CA521 FALA against SARS-CoV-2.
Commun Biol, 4, 2021
|
|
2B34
 
 | | Structure of MAR1 Ribonuclease from Caenorhabditis elegans | | Descriptor: | MAR1 Ribonuclease | | Authors: | Schormann, N, Karpova, E, Li, S, Symersky, J, Zhang, Y, Lu, S, Zhou, Q, Lin, G, Cao, Z, Luo, M, Qiu, S, Luan, C.-H, Luo, D, Huang, W, Shang, Q, McKinstry, A, An, J, Tsao, J, Carson, M, Stinnett, M, Chen, Y, Johnson, D, Gary, R, Arabshahi, A, Bunzel, R, Bray, T, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-09-19 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structure of MAR1 Ribonuclease from Caenorhabditis elegans
To be Published
|
|
5ABT
 
 | | S.enterica HisA mutant D7N, G102A, V106M, D176A | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENE AMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-08 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Protein Structures, Functions and Dynamics of Adaptive Evolution
To be Published
|
|
7FCI
 
 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
