1QJL
 
 | | METALLOTHIONEIN MTA FROM SEA URCHIN (BETA DOMAIN) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
1QJK
 
 | | Metallothionein MTA from sea urchin (alpha domain) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
1AG2
 
 | | PRION PROTEIN DOMAIN PRP(121-231) FROM MOUSE, NMR, 2 MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Billeter, M, Riek, R, Wider, G, Wuthrich, K, Hornemann, S, Glockshuber, R. | | Deposit date: | 1997-03-31 | | Release date: | 1997-10-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the mouse prion protein domain PrP(121-231).
Nature, 382, 1996
|
|
2BEG
 
 | | 3D Structure of Alzheimer's Abeta(1-42) fibrils | | Descriptor: | Amyloid beta A4 protein | | Authors: | Luhrs, T, Ritter, C, Adrian, M, Riek-Loher, D, Bohrmann, B, Dobeli, H, Schubert, D, Riek, R. | | Deposit date: | 2005-10-24 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 3D structure of Alzheimer's amyloid-{beta}(1-42) fibrils.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
9FYP
 
 | | Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
7QTR
 
 | | GB1 in mammalian cells, 50 uM | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Gerez, J.A, Prymaczok, N.C, Kadavath, H, Gosh, D, Butikofer, M, Guntert, P, Riek, R. | | Deposit date: | 2022-01-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in human cells by in-cell NMR and a reporter system to optimize protein delivery or transexpression.
Commun Biol, 5, 2022
|
|
7QTS
 
 | | GB1 in mammalian cells, 10 uM | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Gerez, J.A, Prymaczok, N.C, Kadavath, H, Gosh, D, Butikofer, M, Guntert, P, Riek, R. | | Deposit date: | 2022-01-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in human cells by in-cell NMR and a reporter system to optimize protein delivery or transexpression.
Commun Biol, 5, 2022
|
|
7QCX
 
 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, apo form | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
7QCY
 
 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, complexed with RA-GEF2 peptide | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
8PK2
 
 | | Cryo EM structure of the type 1m polymorph of alpha-synuclein | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
8PIX
 
 | | Cryo EM structure of the type 3C polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
8PJO
 
 | | Cryo EM structure of the type 3D polymorph of alpha-synuclein E46K mutant at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
8PK4
 
 | | Cryo EM structure of the type 5A polymorph of alpha-synuclein. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
8QUT
 
 | |
8QV8
 
 | |
2WVO
 
 | | Structure of the HET-S N-terminal domain | | Descriptor: | CHLORIDE ION, SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Prion Inhibition by Het-S.
Mol.Cell, 38, 2010
|
|
6H6B
 
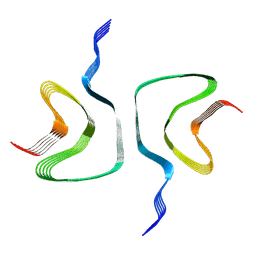 | | Structure of alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Mona, D, Ringler, P, Lauer, M.E, Riek, R, Britschgi, M, Stahlberg, H. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of alpha-synuclein fibrils.
Elife, 7, 2018
|
|
6TUB
 
 | | Beta-endorphin amyloid fibril | | Descriptor: | Beta-endorphin | | Authors: | Verasdonck, J, Seuring, C, Gath, J, Ghosh, D, Nespovitaya, N, Waelti, M.A, Maji, S, Cadalbert, R, Boeckmann, A, Guentert, P, Meier, B.H, Riek, R. | | Deposit date: | 2020-01-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The three-dimensional structure of human beta-endorphin amyloid fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1U34
 
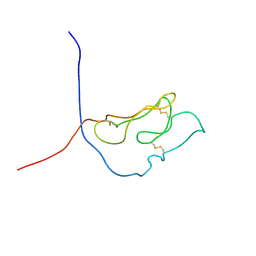 | | 3D NMR structure of the first extracellular domain of CRFR-2beta, a type B1 G-protein coupled receptor | | Descriptor: | Corticotropin releasing factor receptor 2 | | Authors: | Grace, C.R, Perrin, M.H, DiGruccio, M.R, Miller, C.L, Rivier, J.E, Vale, W.W, Riek, R. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure and peptide hormone binding site of the first extracellular domain of a type B1 G protein-coupled receptor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8PXS
 
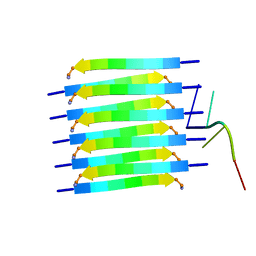 | | Short RNA binding to peptide amyloids | | Descriptor: | RNA (5'-R(P*GP*UP*CP*A)-3'), VAL-ALA-GLN-ALA-GLN-ILE-ASN-ILE | | Authors: | Rout, S.K, Cadalbert, R, Schroder, N, Wiegand, T, Zehnder, J, Gampp, O, Guntert, P, Kringler, D, Kreutz, C, Knorlein, A, Hall, J, Greenwald, J, Riek, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | SOLID-STATE NMR | | Cite: | An Analysis of Nucleotide-Amyloid Interactions Reveals Selective Binding to Codon-Sized RNA.
J.Am.Chem.Soc., 145, 2023
|
|
5LAO
 
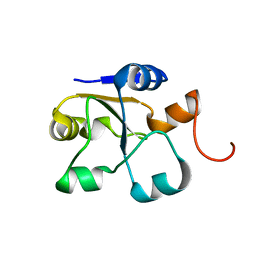 | | S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5LAM
 
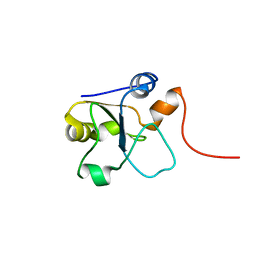 | | Refined 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5OQK
 
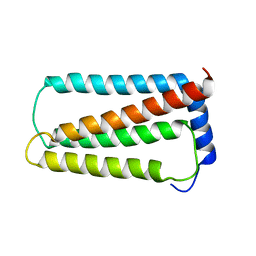 | | Solution NMR structure of truncated, human Hv1/VSOP (Voltage-gated proton channel) | | Descriptor: | Voltage-gated hydrogen channel 1 | | Authors: | Bayrhuber, M, Maslennikov, I, Kwiatowski, W, Sobol, A, Wierschem, C, Eichmann, C, Riek, R. | | Deposit date: | 2017-08-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure and Functional Behavior of the Human Proton Channel.
Biochemistry, 58, 2019
|
|
5C5A
 
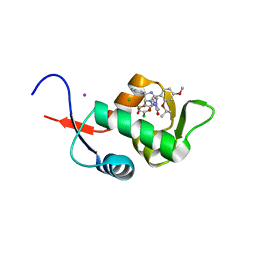 | | Crystal Structure of HDM2 in complex with Nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Orts, J, Waelti, M.A, Marsh, M, Vera, L, Gossert, A.D, Guentert, P, Riek, R. | | Deposit date: | 2015-06-19 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.146 Å) | | Cite: | NMR Molecular Replacement, NMR2
To Be Published
|
|
1LG4
 
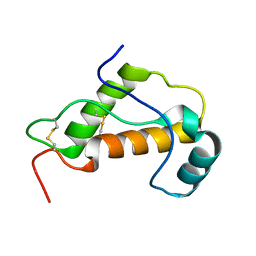 | |
