3DTU
 
 | | Catalytic core subunits (I and II) of cytochrome c oxidase from Rhodobacter sphaeroides complexed with deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CADMIUM ION, CALCIUM ION, ... | | Authors: | Qin, L, Mills, D.A, Buhrow, L, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A conserved steroid binding site in cytochrome C oxidase.
Biochemistry, 47, 2008
|
|
3FYE
 
 | | Catalytic core subunits (I and II) of cytochrome c oxidase from Rhodobacter sphaeroides in the reduced state | | Descriptor: | CADMIUM ION, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Qin, L, Mills, D.A, Proshlyakov, D.A, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Redox dependent conformational changes in cytochrome c oxidase suggest a gating mechanism for proton uptake.
Biochemistry, 48, 2009
|
|
3FYI
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides in the reduced state bound with cyanide | | Descriptor: | CADMIUM ION, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Qin, L, Mills, D.A, Proshlyakov, D.A, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redox dependent conformational changes in cytochrome c oxidase suggest a gating mechanism for proton uptake.
Biochemistry, 48, 2009
|
|
6BG2
 
 | |
6C0U
 
 | | Crystal structure of cAMP-dependent protein kinase Calpha subunit bound with N46 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R,4R)-4-{[4-(2-fluoro-3-methoxy-6-propoxybenzene-1-carbonyl)benzene-1-carbonyl]amino}pyrrolidin-3-yl]-1H-indazole-5-carboxamide, PHOSPHATE ION, ... | | Authors: | Qin, L, Sankaran, B, Kim, C. | | Deposit date: | 2018-01-02 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for selective inhibition of human PKG I alpha by the balanol-like compound N46.
J. Biol. Chem., 293, 2018
|
|
6C0T
 
 | | Crystal structure of cGMP-dependent protein kinase Ialpha (PKG Ialpha) catalytic domain bound with N46 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R,4R)-4-{[4-(2-fluoro-3-methoxy-6-propoxybenzene-1-carbonyl)benzene-1-carbonyl]amino}pyrrolidin-3-yl]-1H-indazole-5-carboxamide, TRIETHYLENE GLYCOL, ... | | Authors: | Qin, L, Sankaran, B, Kim, C. | | Deposit date: | 2018-01-02 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selective inhibition of human PKG I alpha by the balanol-like compound N46.
J. Biol. Chem., 293, 2018
|
|
4RWS
 
 | | Crystal structure of CXCR4 and viral chemokine antagonist vMIP-II complex (PSI Community Target) | | Descriptor: | C-X-C chemokine receptor type 4/Endolysin chimeric protein, Viral macrophage inflammatory protein 2 | | Authors: | Qin, L, Kufareva, I, Holden, L, Wang, C, Zheng, Y, Wu, H, Fenalti, G, Han, G.W, Cherezov, V, Abagyan, R, Stevens, R.C, Handel, T.M, GPCR Network (GPCR) | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine.
Science, 347, 2015
|
|
2GSM
 
 | | Catalytic Core (Subunits I and II) of Cytochrome c oxidase from Rhodobacter sphaeroides | | Descriptor: | CADMIUM ION, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Qin, L, Hiser, C, Mulichak, A, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3PFX
 
 | | Crystal structure of Cel7A from Talaromyces emersonii in complex with cellobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiohydrolase 1 catalytic domain, SULFATE ION, ... | | Authors: | Qin, L, Pereira, J.H, McAndrew, R.P, Simmons, B.A, Sapra, R, Adams, P.D, Sale, K.L. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2616 Å) | | Cite: | Crystal structure of Cel7A from Talaromyces emersonii in complex with cellobiose
TO BE PUBLISHED
|
|
3PFZ
 
 | | Crystal structure of Cel7A from Talaromyces emersonii in complex with cellotetraose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiohydrolase 1 catalytic domain, SULFATE ION, ... | | Authors: | Qin, L, Pereira, J.H, McAndrew, R.P, Simmons, B.A, Sapra, R, Adams, P.D, Sale, K.L. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.0993 Å) | | Cite: | Crystal structure of Cel7A from Talaromyces emersonii in complex with cellotetraose
TO BE PUBLISHED
|
|
5COA
 
 | | Crystal structure of iridoid synthase at 2.2-angstrom resolution | | Descriptor: | HEXAETHYLENE GLYCOL, Iridoid synthase, SULFATE ION | | Authors: | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
5COB
 
 | | Crystal structure of iridoid synthase in complex with NADP+ and 8-oxogeranial at 2.65-angstrom resolution | | Descriptor: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, Iridoid synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
6BDL
 
 | |
7W6K
 
 | | Cryo-EM structure of GmALMT12/QUAC1 anion channel | | Descriptor: | GmALMT12/QUAC1 | | Authors: | Qin, L, Tang, L.H, Xu, J.S, Zhang, X.H, Zhu, Y, Sun, F, Su, M, Zhai, Y.J, Chen, Y.H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure and electrophysiological characterization of ALMT from Glycine max reveal a previously uncharacterized class of anion channels.
Sci Adv, 8, 2022
|
|
7L4U
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 1h | | Descriptor: | (5S)-5-(3-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidin-1-yl}-3-oxopropyl)pyrrolidin-2-one, CHLORIDE ION, Monoglyceride lipase | | Authors: | Qin, L, Lane, W, Skene, R.J, Dougan, D. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L4W
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 2d | | Descriptor: | (2s,4R)-2-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidine-1-carbonyl}-7-oxa-5-azaspiro[3.4]octan-6-one, Monoglyceride lipase | | Authors: | Qin, L, Gay, S.C, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L50
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 4f | | Descriptor: | (2s,4R)-2-{3-[(3-chloro-4-methylphenyl)methoxy]azetidine-1-carbonyl}-7-oxa-5-azaspiro[3.4]octan-6-one, ACETATE ION, Monoglyceride lipase | | Authors: | Qin, L, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L4T
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidine-1-carbonyl}-2H-1,4-benzoxazin-3(4H)-one, ACETATE ION, ... | | Authors: | Qin, L, Gay, S.C, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
5URK
 
 | | Crystal structure of human BRR2 in complex with T-3935799 | | Descriptor: | 6-benzyl-3-[3-(benzyloxy)phenyl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(1H,3H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Qin, L, Tjhen, R, Klein, M.G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
4R4L
 
 | | Crystal structure of wt cGMP dependent protein kinase I alpha (PKGI alpha) leucine zipper | | Descriptor: | HEXANE-1,6-DIOL, SULFATE ION, cGMP-dependent protein kinase 1 | | Authors: | Reger, A.S, Guo, E, Yang, M.P, Qin, L, Kim, C. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structures of cGMP-Dependent Protein Kinase (PKG) I alpha Leucine Zippers Reveal an Interchain Disulfide Bond Important for Dimer Stability.
Biochemistry, 54, 2015
|
|
4R4M
 
 | | Crystal structure of C42L cGMP dependent protein kinase I alpha (PKGI alpha) leucine zipper | | Descriptor: | SULFATE ION, cGMP-dependent protein kinase 1 | | Authors: | Reger, A.S, Guo, E, Yang, M.P, Qin, L, Kim, C. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structures of cGMP-Dependent Protein Kinase (PKG) I alpha Leucine Zippers Reveal an Interchain Disulfide Bond Important for Dimer Stability.
Biochemistry, 54, 2015
|
|
8HF2
 
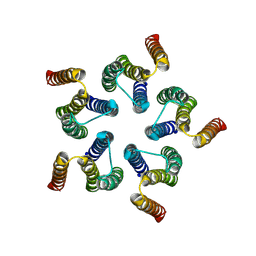 | | Cryo-EM structure of WeiTsing | | Descriptor: | PRA1 family protein | | Authors: | Qin, L, Tang, L.H, Chen, Y.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | WeiTsing, a pericycle-expressed ion channel, safeguards the stele to confer clubroot resistance.
Cell, 186, 2023
|
|
3OGG
 
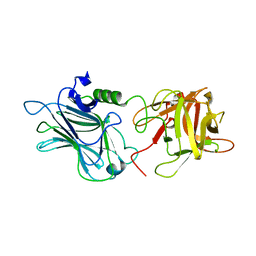 | | Crystal structure of the receptor binding domain of botulinum neurotoxin D | | Descriptor: | Botulinum neurotoxin type D | | Authors: | Zhang, Y, Gao, X, Qin, L, Buchko, G.W, Robinson, H, Varnum, S.M. | | Deposit date: | 2010-08-16 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural analysis of the receptor binding domain of botulinum neurotoxin serotype D.
Biochem.Biophys.Res.Commun., 401, 2010
|
|
8B9F
 
 | | Structure of Echovirus 11 complexed with DAF (CD55) calculated from symmetry expansion | | Descriptor: | Complement decay-accelerating factor, Genome polyprotein, SPHINGOSINE | | Authors: | Stuart, D.I, Ren, J, Qin, L, Zhou, D. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
8B8R
 
 | | Complex of Echovirus 11 with its attaching receptor decay-accelerating factor (CD55) | | Descriptor: | DECAY ACCELERATING FACTOR (CD55), SPHINGOSINE, VP1, ... | | Authors: | Stuart, D.I, Ren, J, Zhou, D, Qin, L. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-07 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
