8JMM
 
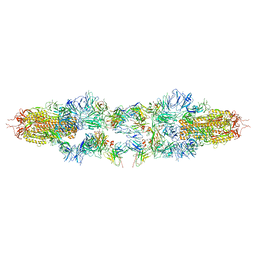 | | Structure of XBB spike protein (S) dimer-trimer in complex with bispecific antibody G7-Fc at 3.75 Angstroms resolution. | | Descriptor: | 7F3 Fv, GW01 Fv, Spike glycoprotein | | Authors: | Hao, A, Mao, Q, Chen, Z, Huang, J, Sun, L. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of XBB spike protein (S) dimer-trimer in complex with bispecific antibody G7-Fc at 3.75 Angstroms resolution.
To Be Published
|
|
8TZK
 
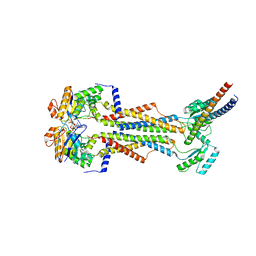 | | Cryo-EM structure of Vibrio cholerae FtsE/FtsX/EnvC complex, shortened | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural insights into the FtsEX-EnvC complex regulation on septal peptidoglycan hydrolysis in Vibrio cholerae.
Structure, 32, 2024
|
|
8TZJ
 
 | | Cryo-EM structure of Vibrio cholerae FtsE/FtsX complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural insights into the FtsEX-EnvC complex regulation on septal peptidoglycan hydrolysis in Vibrio cholerae.
Structure, 32, 2024
|
|
8TZL
 
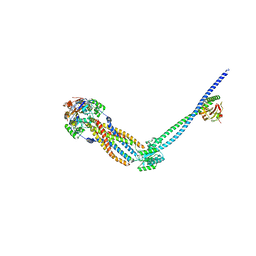 | | Cryo-EM structure of Vibrio cholerae FtsE/FtsX/EnvC complex, full-length | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural insights into the FtsEX-EnvC complex regulation on septal peptidoglycan hydrolysis in Vibrio cholerae.
Structure, 32, 2024
|
|
1MRN
 
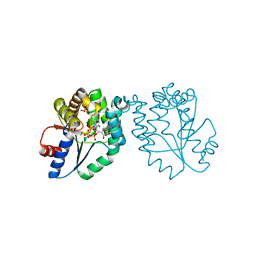 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH BISUBSTRATE INHIBITOR (TP5A) | | Descriptor: | MAGNESIUM ION, P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SULFATE ION, ... | | Authors: | Haouz, A, Vanheusden, V, Munier-Lehmann, H, Froeyen, M, Herdewijn, P, Van Calenbergh, S, Delarue, M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism.
J.Biol.Chem., 278, 2003
|
|
6PLE
 
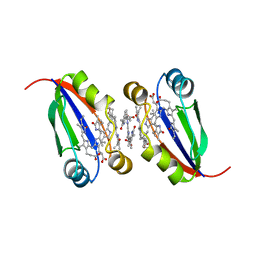 | |
1MRS
 
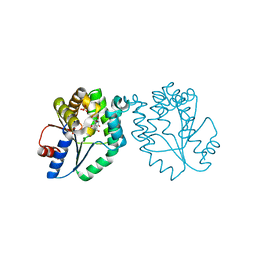 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH 5-CH2OH DEOXYURIDINE MONOPHOSPHATE | | Descriptor: | 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Haouz, A, Vanheusden, V, Munier-Lehmann, H, Froeyen, M, Herdewijn, P, Van Calenbergh, S, Delarue, M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism.
J.Biol.Chem., 278, 2003
|
|
5YJL
 
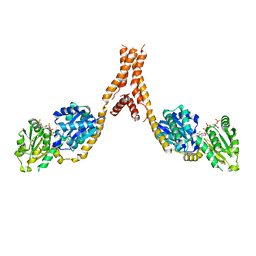 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with NADPH and GBP | | Descriptor: | Glutamyl-tRNA reductase 1, Glutamyl-tRNA reductase-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, A, Han, F. | | Deposit date: | 2017-10-11 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Arabidopsis thaliana glutamyl-tRNAGlureductase in complex with NADPH and glutamyl-tRNAGlureductase binding protein
Photosyn. Res., 137, 2018
|
|
4N7R
 
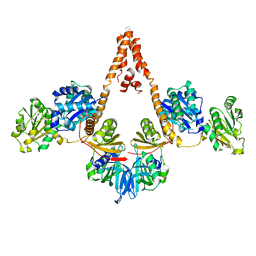 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its binding protein | | Descriptor: | Genomic DNA, chromosome 3, P1 clone: MXL8, ... | | Authors: | Zhao, A, Fang, Y, Lin, Y, Gong, W, Liu, L. | | Deposit date: | 2013-10-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6DS8
 
 | |
6DS7
 
 | |
9B70
 
 | | Cryo-EM structure of MraY in complex with analogue 2 | | Descriptor: | (2~{S},3~{S})-3-[(2~{S},3~{R},4~{S},5~{R})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-2-[[3-[[[(2~{S})-6-azanyl-2-(hexadecanoylamino)hexanoyl]amino]methyl]phenyl]methylamino]-3-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propanoic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2024-03-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Development of a natural product optimization strategy for inhibitors against MraY, a promising antibacterial target.
Nat Commun, 15, 2024
|
|
9B71
 
 | | Cryo-EM structure of MraY in complex with analogue 3 | | Descriptor: | (2~{S},3~{S})-3-[(2~{S},3~{R},4~{S},5~{R})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-3-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]-2-[[4-[[[(2~{S})-5-carbamimidamido-2-(hexadecanoylamino)pentanoyl]amino]methyl]phenyl]methylamino]propanoic acid, MraYAA Nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2024-03-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Development of a natural product optimization strategy for inhibitors against MraY, a promising antibacterial target.
Nat Commun, 15, 2024
|
|
7UQ0
 
 | | Putative periplasmic iron siderophore binding protein FecB (Rv3044) from Mycobacterium tuberculosis | | Descriptor: | CITRIC ACID, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Chao, A, Cuthbert, B.J, Goulding, C.W. | | Deposit date: | 2022-04-18 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differentiating the roles of Mycobacterium tuberculosis substrate binding proteins, FecB and FecB2, in iron uptake.
Plos Pathog., 19, 2023
|
|
3PPD
 
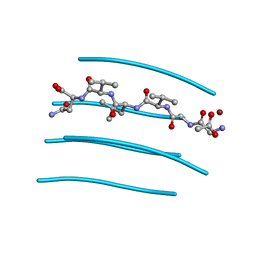 | | GGVLVN segment from Human Prostatic Acid Phosphatase Residues 260-265, involved in Semen-Derived Enhancer of Viral Infection | | Descriptor: | ACETIC ACID, GGVLVN peptide, amyloid forming segment, ... | | Authors: | Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation.
Nature, 475, 2011
|
|
8BVE
 
 | | MoeA2 from Corynebacterium glutamicum | | Descriptor: | CITRIC ACID, Molybdopterin molybdenumtransferase, SODIUM ION | | Authors: | Martinez, M, Haouz, A, Wehenkel, A.M, Alzari, P.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation.
Nat Microbiol, 8, 2023
|
|
8BVF
 
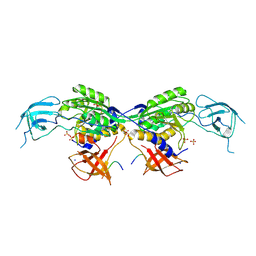 | | MoeA2 from Corynebacterium glutamicum in complex with FtsZ-CTD | | Descriptor: | Cell division protein FtsZ, Molybdopterin molybdenumtransferase, SODIUM ION, ... | | Authors: | Martinez, M, Haouz, A, Wehenkel, A.M, Alzari, P.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation.
Nat Microbiol, 8, 2023
|
|
4USS
 
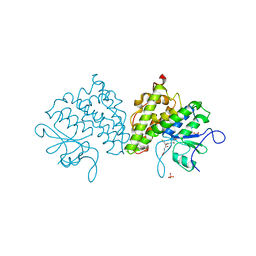 | | Populus trichocarpa glutathione transferase X1-1 (GHR1), complexed with glutathione | | Descriptor: | GLUTATHIONE, GLUTATHIONYL HYDROQUINONE REDUCTASE, PHOSPHATE ION | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Dumaracay, S, Favier, F, Didierjean, C, Saul, F, Haouz, A, Morel-Rouhier, M, Gelhaye, E, Rouhier, N, Hecker, A. | | Deposit date: | 2014-07-13 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutathionyl-Hydroquinone Reductases from Poplar are Plastidial Proteins that Deglutathionylate Both Reduced and Oxidized Glutathionylated Quinones.
FEBS Lett., 589, 2015
|
|
8QKE
 
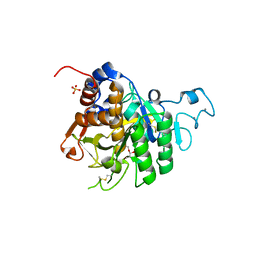 | | PvSub1 Catalytic Domain in Complex with Peptidomimetic Inhibitor (MH-13) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Peptidomimetic Inhibitor (MH-13), ... | | Authors: | Batista, F.A, Martinez, M, Bouillon, A, Mechaly, A, Alzari, P.M, Haouz, A, Barale, J.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Insights from structure-activity relationships and the binding mode of peptidic alpha-ketoamide inhibitors of the malaria drug target subtilisin-like SUB1.
Eur.J.Med.Chem., 269, 2024
|
|
8QKJ
 
 | | PvSub1 Catalytic Domain in Complex with Peptidomimetic Inhibitor (MAM-133) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Peptidomimetic Inhibitor (MAM-133), ... | | Authors: | Batista, F.A, Martinez, M, Bouillon, A, Mechaly, A, Alzari, P.M, Haouz, A, Barale, J.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Insights from structure-activity relationships and the binding mode of peptidic alpha-ketoamide inhibitors of the malaria drug target subtilisin-like SUB1.
Eur.J.Med.Chem., 269, 2024
|
|
8QKG
 
 | | PvSub1 Catalytic Domain in Complex with Peptidomimetic Inhibitor (MAM-125) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Peptidomimetic Inhibitor (MAM-125), ... | | Authors: | Batista, F.A, Martinez, M, Bouillon, A, Mechaly, A, Alzari, P.M, Haouz, A, Barale, J.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Insights from structure-activity relationships and the binding mode of peptidic alpha-ketoamide inhibitors of the malaria drug target subtilisin-like SUB1.
Eur.J.Med.Chem., 269, 2024
|
|
3NFK
 
 | | Crystal structure of the PTPN4 PDZ domain complexed with the C-terminus of a rabies virus G protein | | Descriptor: | GLYCEROL, Glycoprotein G, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Babault, N, Cordier, F, Lafage, M, Cockburn, J, Haouz, A, Rey, F.A, Delepierre, M, Buc, H, Lafon, M, Wolff, N. | | Deposit date: | 2010-06-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Peptides Targeting the PDZ Domain of PTPN4 Are Efficient Inducers of Glioblastoma Cell Death.
Structure, 19, 2011
|
|
3NFL
 
 | | Crystal structure of the PTPN4 PDZ domain complexed with the C-terminus of the GluN2A NMDA receptor subunit | | Descriptor: | Glutamate [NMDA] receptor subunit epsilon-1, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Babault, N, Cordier, F, Lafage, M, Cockburn, J, Haouz, A, Rey, F.A, Delepierre, M, Buc, H, Lafon, M, Wolff, N. | | Deposit date: | 2010-06-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Peptides Targeting the PDZ Domain of PTPN4 Are Efficient Inducers of Glioblastoma Cell Death.
Structure, 19, 2011
|
|
3N8D
 
 | | Crystal structure of Staphylococcus aureus VRSA-9 D-Ala:D-Ala ligase | | Descriptor: | CHLORIDE ION, D-alanine--D-alanine ligase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Saul, F.A, Haouz, A, Meziane-Cherif, D. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of vancomycin dependence in VanA-type Staphylococcus aureus VRSA-9.
J.Bacteriol., 192, 2010
|
|
8Q5P
 
 | | Structure of the lysine methyltransferase SETD2 in complex with a peptide derived from human tyrosine kinase ACK1 | | Descriptor: | Activated CDC42 kinase 1, Histone-lysine N-methyltransferase SETD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Mechaly, A, Le Coadou, L, Dupret, J.M, Haouz, A, Rodrigues Lima, F. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Structural and enzymatic evidence for the methylation of the ACK1 tyrosine kinase by the histone lysine methyltransferase SETD2.
Biochem.Biophys.Res.Commun., 695, 2024
|
|
