1ATJ
 
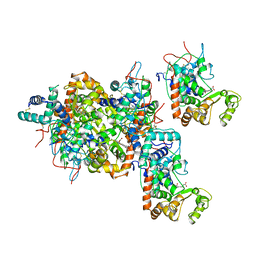 | | RECOMBINANT HORSERADISH PEROXIDASE C1A | | Descriptor: | CALCIUM ION, PEROXIDASE C1A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gajhede, M, Schuller, D.J, Henriksen, A, Smith, A.T, Poulos, T.L. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of horseradish peroxidase C at 2.15 A resolution.
Nat.Struct.Biol., 4, 1997
|
|
1PSP
 
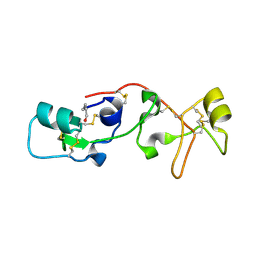 | | PANCREATIC SPASMOLYTIC POLYPEPTIDE: FIRST THREE-DIMENSIONAL STRUCTURE OF A MEMBER OF THE MAMMALIAN TREFOIL FAMILY OF PEPTIDES | | Descriptor: | PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | Gajhede, M, Petersen, T.N, Henriksen, A, Petersen, J.F.W, Dauter, Z, Wilson, K.S, Thim, L. | | Deposit date: | 1994-01-05 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pancreatic spasmolytic polypeptide: first three-dimensional structure of a member of the mammalian trefoil family of peptides.
Structure, 1, 1993
|
|
1BV1
 
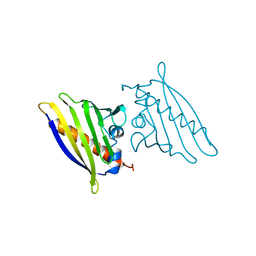 | | BIRCH POLLEN ALLERGEN BET V 1 | | Descriptor: | BET V 1 | | Authors: | Gajhede, M, Osmark, P, Poulsen, F.M, Ipsen, H, Larson, J.N, Joostvan, R.J, Schou, C, Lowenstein, H, Spangfort, M.D. | | Deposit date: | 1997-07-08 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
1BTV
 
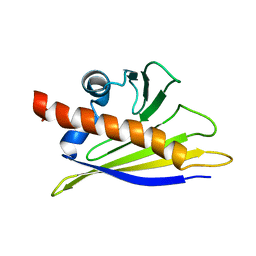 | | STRUCTURE OF BET V 1, NMR, 20 STRUCTURES | | Descriptor: | BET V 1 | | Authors: | Osmark, P, Poulsen, F.M, Gajhede, M, Larsen, J.N, Spangfort, M.D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
7ATJ
 
 | | RECOMBINANT HORSERADISH PEROXIDASE C1A COMPLEX WITH CYANIDE AND FERULIC ACID | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CALCIUM ION, CYANIDE ION, ... | | Authors: | Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 1999-04-26 | | Release date: | 2000-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates.
J.Biol.Chem., 274, 1999
|
|
6ATJ
 
 | | RECOMBINANT HORSERADISH PEROXIDASE C COMPLEX WITH FERULIC ACID | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 1999-04-23 | | Release date: | 2000-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates.
J.Biol.Chem., 274, 1999
|
|
2ANJ
 
 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
2PSP
 
 | |
1GWO
 
 | |
1GX2
 
 | |
3BFU
 
 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | Descriptor: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
4MH5
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
9ERX
 
 | | Structural basis of D9-THC analog activity at the Cannabinoid 1 receptor | | Descriptor: | (6aR,10aR)-9-(hydroxymethyl)-6,6-dimethyl-3-(2-methyloctan-2-yl)-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, Antibody ScFv16 Fab fragment, Cannabinoid receptor 1, ... | | Authors: | Thorsen, T.S, Kulkarni, Y, Boggild, A, Drace, T, Nissen, P, Gajhede, M, Boesen, T, Kastrup, J.S, Gloriam, D. | | Deposit date: | 2024-03-25 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of Delta 9 -THC analog activity at the Cannabinoid 1 receptor.
Res Sq, 2024
|
|
5OY3
 
 | | The structural basis of the histone demethylase KDM6B histone 3 lysine 27 specificity | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (III) ION, ... | | Authors: | Jones, S.E, Olsen, L, Gajhede, M. | | Deposit date: | 2017-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.136 Å) | | Cite: | Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity.
Biochemistry, 57, 2018
|
|
5CC2
 
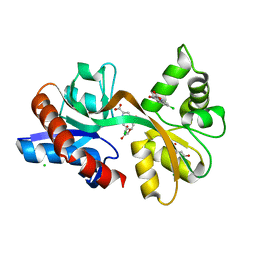 | | STRUCTURE OF THE LIGAND-BINDING DOMAIN OF THE IONOTROPIC GLUTAMATE RECEPTOR-LIKE GLUD2 IN COMPLEX WITH 7-CKA | | Descriptor: | 7-Chlorokynurenic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Pharmacology and Structural Analysis of Ligand Binding to the Orthosteric Site of Glutamate-Like GluD2 Receptors.
Mol.Pharmacol., 89, 2016
|
|
2ATJ
 
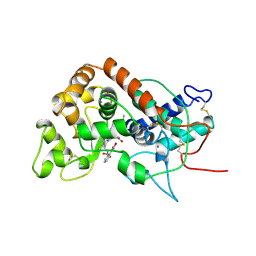 | | RECOMBINANT HORSERADISH PEROXIDASE COMPLEX WITH BENZHYDROXAMIC ACID | | Descriptor: | BENZHYDROXAMIC ACID, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Henriksen, A, Schuller, D.J, Gajhede, M. | | Deposit date: | 1997-08-19 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural interactions between horseradish peroxidase C and the substrate benzhydroxamic acid determined by X-ray crystallography.
Biochemistry, 37, 1998
|
|
5JT8
 
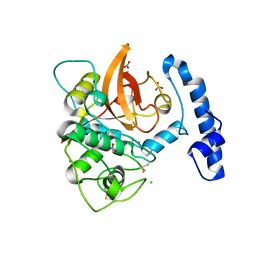 | | Structural basis for the limited antibody cross reactivity between the mite allergens Blo t 1 and Der p 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Blo t 1 allergen, ... | | Authors: | Meno, K, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the mite allergen Blo t 1 explains the limited antibody cross-reactivity to Der p 1.
Allergy, 72, 2017
|
|
1KZM
 
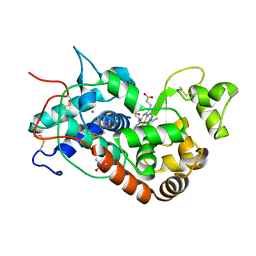 | | Distal Heme Pocket Mutant (R38S/H42E) of Recombinant Horseradish Peroxidase C (HRP C). | | Descriptor: | CACODYLATE ION, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Meno, K, Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 2002-02-07 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the two horseradish peroxidase catalytic residue variants H42E and R38S/H42E: implications for the catalytic cycle.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2PBW
 
 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex with the Partial agonist Domoic Acid at 2.5 A Resolution | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, Glutamate receptor, ionotropic kainate 1 | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
4ATJ
 
 | | DISTAL HEME POCKET MUTANT (H42E) OF RECOMBINANT HORSERADISH PEROXIDASE IN COMPLEX WITH BENZHYDROXAMIC ACID | | Descriptor: | BENZHYDROXAMIC ACID, CALCIUM ION, PROTEIN (PEROXIDASE C1A), ... | | Authors: | Meno, K, Jennings, S, Smith, A.T, Henriksen, A, Gajhede, M. | | Deposit date: | 1999-04-19 | | Release date: | 2002-10-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the two horseradish peroxidase catalytic residue variants H42E and R38S/H42E: implications for the catalytic cycle.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6F6D
 
 | | The catalytic domain of KDM6B in complex with H3(17-33)K18IA21M peptide | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Histone 3 peptide H3(17-33)K18IA21M, ... | | Authors: | Jones, S.E, Olsen, L, Gajhede, M. | | Deposit date: | 2017-12-05 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81810677 Å) | | Cite: | Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity.
Biochemistry, 57, 2018
|
|
6RUQ
 
 | | Structure of GluA2cryst in complex the antagonist ZK200775 and the negative allosteric modulator GYKI53655 at 4.65 A resolution | | Descriptor: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, Glutamate receptor 2, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Krintel, C, Venskutonyte, R, Mirza, O.A, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.65 Å) | | Cite: | Binding of a negative allosteric modulator and competitive antagonist can occur simultaneously at the ionotropic glutamate receptor GluA2.
Febs J., 288, 2021
|
|
1QO4
 
 | | ARABIDOPSIS THALIANA PEROXIDASE A2 AT ROOM TEMPERATURE | | Descriptor: | CALCIUM ION, PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Henriksen, A, Gajhede, M. | | Deposit date: | 1999-11-02 | | Release date: | 2000-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential Activity and Structure of Highly Similar Peroxidases. Spectroscopic, Crystallographic, and Enzymatic Analyses of Lignifying Arabidopsis Thaliana Peroxidase A2 and Horseradish Peroxidase A2
Biochemistry, 40, 2001
|
|
1S46
 
 | | Covalent intermediate of the E328Q amylosucrase mutant | | Descriptor: | amylosucrase, beta-D-glucopyranose | | Authors: | Jensen, M.H, Mirza, O, Albenne, C, Remaud-Simeon, M, Monsan, P, Gajhede, M, Skov, L.K. | | Deposit date: | 2004-01-15 | | Release date: | 2004-03-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the covalent intermediate of amylosucrase from Neisseria polysaccharea.
Biochemistry, 43, 2004
|
|
1QNX
 
 | | Ves v 5, an allergen from Vespula vulgaris venom | | Descriptor: | SODIUM ION, VES V 5 | | Authors: | Henriksen, A, Gajhede, M, Spangfort, M.D. | | Deposit date: | 1999-10-25 | | Release date: | 2000-10-26 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Major Venom Allergen of Yellow Jackets, Ves V 5: Structural Characterization of a Pathogenesis-Related Protein Superfamily.
Proteins: Struct.,Funct., Genet., 45, 2001
|
|
