6KCR
 
 | |
6MTY
 
 | | Crystal structure of a human anti-ZIKV-DENV neutralizing antibody MZ4 isolated following ZPIV vaccination | | Descriptor: | MZ4 Heavy Chain, MZ4 Light Chain | | Authors: | Sankhala, R.S, Dussupt, V, Donofrio, G, Choe, M, Modjarrad, K, Michael, N.L, Krebs, S.J, Joyce, M.G. | | Deposit date: | 2018-10-22 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor.
Nat Med, 26, 2020
|
|
6MTX
 
 | | Crystal structure of a human anti-ZIKV-DENV neutralizing antibody MZ1 isolated following ZPIV vaccination | | Descriptor: | GLYCEROL, MZ1 Heavy Chain, MZ1 Light Chain | | Authors: | Sankhala, R.S, Dussupt, V, Donofrio, G, Choe, M, Modjarrad, K, Michael, N.L, Krebs, S.J, Joyce, M.G. | | Deposit date: | 2018-10-22 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor.
Nat Med, 26, 2020
|
|
6NIP
 
 | | Crystal structure of a human anti-ZIKV-DENV neutralizing antibody MZ1 in complex with ZIKV E glycoprotein | | Descriptor: | Envelope protein E, MZ1 Heavy chain, MZ1 Light Chain | | Authors: | Sankhala, R.S, Dussupt, V, Donofrio, G, Choe, M, Modjarrad, K, Michael, N.L, Krebs, S.J, Joyce, M.G. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.16 Å) | | Cite: | Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor.
Nat Med, 26, 2020
|
|
6NIU
 
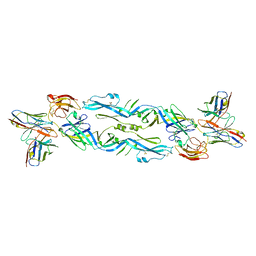 | | Crystal structure of a human anti-ZIKV-DENV neutralizing antibody MZ4 in complex with ZIKV E glycoprotein | | Descriptor: | Envelope protein E, Human MZ4 Fab heavy chain, Human MZ4 Fab light chain | | Authors: | Sankhala, R.S, Dussupt, V, Donofrio, G, Choe, M, Modjarrad, K, Michael, N.L, Krebs, S.J, Joyce, M.G. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.303 Å) | | Cite: | Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor.
Nat. Med., 26, 2020
|
|
6NIS
 
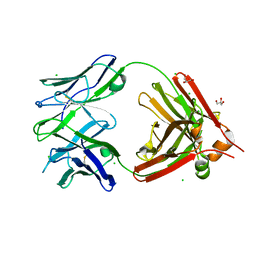 | | Crystal structure of a human anti-ZIKV-DENV neutralizing antibody MZ24 isolated following ZPIV vaccination | | Descriptor: | CHLORIDE ION, GLYCEROL, MZ24 antibody heavy chain, ... | | Authors: | Sankhala, R.S, Dussupt, V, Donofrio, G, Choe, M, Modjarrad, K, Michael, N.L, Krebs, S.J, Joyce, M.G. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor.
Nat Med, 26, 2020
|
|
6U6O
 
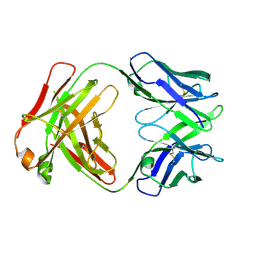 | | Crystal structure of a vaccine-elicited anti-HIV-1 rhesus macaque antibody DH846 | | Descriptor: | DH846 Fab heavy chain, DH846 Fab light chain | | Authors: | Chen, W.-H, Choe, M, Saunders, K.O, Joyce, M.G. | | Deposit date: | 2019-08-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural and genetic convergence of HIV-1 neutralizing antibodies in vaccinated non-human primates.
Plos Pathog., 17, 2021
|
|
6U6M
 
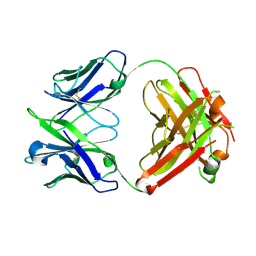 | | Crystal structure of a vaccine-elicited anti-HIV-1 rhesus macaque antibody DH840.1 | | Descriptor: | DH840.1 Fab heavy chain, DH840.1 Fab light chain | | Authors: | Chen, W.-H, Choe, M, Saunders, K.O, Joyce, M.G. | | Deposit date: | 2019-08-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | Structural and genetic convergence of HIV-1 neutralizing antibodies in vaccinated non-human primates.
Plos Pathog., 17, 2021
|
|
4NIX
 
 | | Crystal structure of trypsiligase (K60E/N143H/Y151H/D189K trypsin) orthorhombic form, zinc-bound | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Schoepfel, M, Parthier, C, Stubbs, M.T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | N-terminal protein modification by substrate-activated reverse proteolysis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NIW
 
 | |
4NIV
 
 | |
4NIY
 
 | | Crystal structure of trypsiligase (K60E/N143H/Y151H/D189K trypsin) complexed to YRH-ecotin (M84Y/M85R/A86H ecotin) | | Descriptor: | CALCIUM ION, Cationic trypsin, Ecotin, ... | | Authors: | Schoepfel, M, Parthier, C, Stubbs, M.T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | N-terminal protein modification by substrate-activated reverse proteolysis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3QK1
 
 | | Crystal Structure of Enterokinase-like Trypsin Variant | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Schoepfel, M, Tziridis, A, Arnold, U, Stubbs, M.T. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Towards a restriction proteinase: construction of a self-activating enzyme.
Chembiochem, 12, 2011
|
|
6VBP
 
 | |
6VBQ
 
 | |
6VBO
 
 | |
6C6X
 
 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque. | | Descriptor: | JC57-14 Heavy chain, JC57-14 Light chain | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
6C6Z
 
 | |
7U8E
 
 | |
5WHZ
 
 | | PGDM1400-10E8v4 CODV Fab | | Descriptor: | Anti-HIV CODV-Fab Heavy chain, Anti-HIV CODV-Fab Light chain | | Authors: | Lord, D.M, Wei, R.R. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.549 Å) | | Cite: | Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques.
Science, 358, 2017
|
|
5WDF
 
 | |
8SMT
 
 | | Crystal structure of antibody WRAIR-2134 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2134 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8SMI
 
 | | Crystal structure of antibody WRAIR-2123 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2123 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8SGU
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-13 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
6C6Y
 
 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque in complex with MERS Receptor Binding Domain | | Descriptor: | JC57-14 Heavy chain, JC57-14 Light chain, SULFATE ION, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
