1C53
 
 | | S-CLASS CYTOCHROMES C HAVE A VARIETY OF FOLDING PATTERNS: STRUCTURE OF CYTOCHROME C-553 FROM DESULFOVIBRIO VULGARIS DETERMINED BY THE MULTI-WAVELENGTH ANOMALOUS DISPERSION METHOD | | Descriptor: | CYTOCHROME C553, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakagawa, A, Higuchi, Y, Yasuoka, N, Katsube, Y, Yaga, T. | | Deposit date: | 1991-08-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-class cytochromes c have a variety of folding patterns: structure of cytochrome c-553 from Desulfovibrio vulgaris determined by the multi-wavelength anomalous dispersion method.
J.Biochem.(Tokyo), 108, 1990
|
|
1BK7
 
 | | RIBONUCLEASE MC1 FROM THE SEEDS OF BITTER GOURD | | Descriptor: | PROTEIN (RIBONUCLEASE MC1) | | Authors: | Nakagawa, A, Tanaka, I. | | Deposit date: | 1998-07-15 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a ribonuclease from the seeds of bitter gourd (Momordica charantia) at 1.75 A resolution.
Biochim.Biophys.Acta, 1433, 1999
|
|
1UF2
 
 | | The Atomic Structure of Rice dwarf Virus (RDV) | | Descriptor: | Core protein P3, Outer capsid protein P8, Structural protein P7 | | Authors: | Nakagawa, A, Miyazaki, N, Taka, J, Naitow, H, Ogawa, A, Fujimoto, Z, Mizuno, H, Higashi, T, Watanabe, Y, Omura, T, Cheng, R.H, Tsukihara, T. | | Deposit date: | 2003-05-23 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins.
Structure, 11, 2003
|
|
1RL2
 
 | |
4NI0
 
 | | Quaternary R3 CO-liganded hemoglobin structure in complex with a thiol containing compound | | Descriptor: | 5-[(2S)-2,3-dihydro-1,4-benzodioxin-2-yl]-2,4-dihydro-3H-1,2,4-triazole-3-thione, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Safo, M.K, Meadows, J, Ko, T.-P, Nakagawa, A, Zapol, W. | | Deposit date: | 2013-11-05 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of a Small Molecule that Increases Hemoglobin Oxygen Affinity and Reduces SS Erythrocyte Sickling.
Acs Chem.Biol., 9, 2014
|
|
4NI1
 
 | | Quaternary R CO-liganded hemoglobin structure in complex with a thiol containing compound | | Descriptor: | 5-[(2R)-2,3-dihydro-1,4-benzodioxin-2-yl]-2,4-dihydro-3H-1,2,4-triazole-3-thione, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Safo, M.K, Meadows, J, Ko, T.-P, Nakagawa, A, Zapol, W. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a Small Molecule that Increases Hemoglobin Oxygen Affinity and Reduces SS Erythrocyte Sickling.
Acs Chem.Biol., 9, 2014
|
|
1F2D
 
 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Yao, M, Ose, T, Sugimoto, H, Horiuchi, A, Nakagawa, A, Yokoi, D, Murakami, T, Honma, M, Wakatsuki, S, Tanaka, I. | | Deposit date: | 2000-05-24 | | Release date: | 2000-12-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus.
J.Biol.Chem., 275, 2000
|
|
1MR8
 
 | | MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 FROM HUMAN | | Descriptor: | CALCIUM ION, MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 | | Authors: | Ishikawa, K, Nakagawa, A, Tanaka, I, Nishihira, J. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of human MRP8, a member of the S100 calcium-binding protein family, by MAD phasing at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5X70
 
 | |
5X6X
 
 | |
3KLR
 
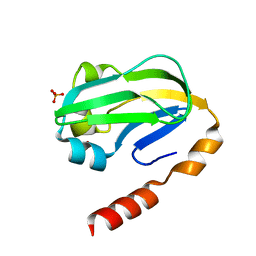 | | Bovine H-protein at 0.88 angstrom resolution | | Descriptor: | GLYCEROL, Glycine cleavage system H protein, SULFATE ION | | Authors: | Higashiura, A, Kurakane, T, Matsuda, M, Suzuki, M, Inaka, K, Sato, M, Tanaka, H, Fujiwara, K, Nakagawa, A. | | Deposit date: | 2009-11-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | High-resolution X-ray crystal structure of bovine H-protein at 0.88 A resolution
Acta Crystallogr.,Sect.D, 66, 2010
|
|
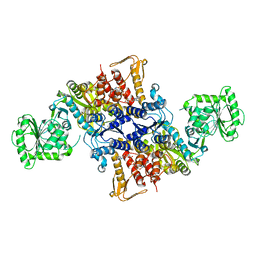
5X6Z
 
 | |
5X6Y
 
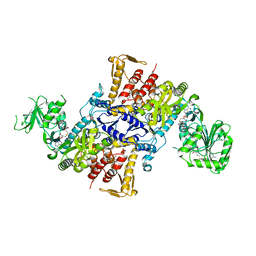 | |
4WY3
 
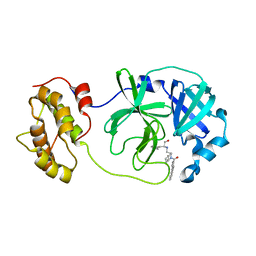 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (R,S)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3R,4aS,8aR)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-11-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
8HEW
 
 | | Potato 14-3-3 St14f | | Descriptor: | 14-3-3 protein, StFDL1 peptide | | Authors: | Taoka, K, Kawahara, I, Shinya, S, Harada, K, Muranaka, T, Furuita, K, Nakagawa, A, Fujiwara, T, Tsuji, H, Kojima, C. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Multifunctional chemical inhibitors of the florigen activation complex discovered by structure-based high-throughput screening.
Plant J., 112, 2022
|
|
5H5L
 
 | | Structure of prostaglandin synthase D of Nilaparvata lugens | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, S, Nakagawa, A. | | Deposit date: | 2016-11-07 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular structure of a prostaglandin D synthase requiring glutathione from the brown planthopper, Nilaparvata lugens
Biochem. Biophys. Res. Commun., 492, 2017
|
|
4TWW
 
 | | Structure of SARS-3CL protease complex with a Bromobenzoyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3S,4aR,8aS)-2-(4-bromobenzoyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4TWY
 
 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3S,4aR,8aS)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4TRY
 
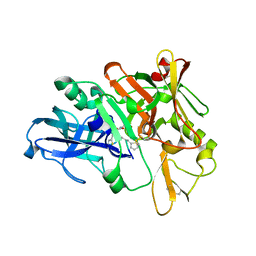 | | Structure of BACE1 complex with a HEA-type inhibitor | | Descriptor: | Beta-secretase 1, GLU-ILE-TIH-THC-NVA | | Authors: | Akaji, K, Teruya, K, Akiyama, T, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of BACE1 complex with an anti-HMC-type inhibitor
to be published
|
|
1FIM
 
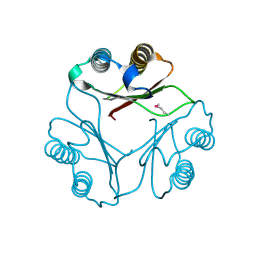 | |
1HUS
 
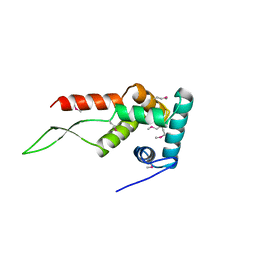 | | RIBOSOMAL PROTEIN S7 | | Descriptor: | RIBOSOMAL PROTEIN S7 | | Authors: | Hosaka, H, Nakagawa, A, Tanaka, I. | | Deposit date: | 1997-08-08 | | Release date: | 1998-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ribosomal protein S7: a new RNA-binding motif with structural similarities to a DNA architectural factor.
Structure, 5, 1997
|
|
1WP1
 
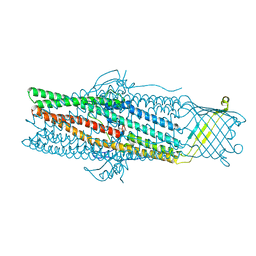 | | Crystal structure of the drug-discharge outer membrane protein, OprM | | Descriptor: | Outer membrane protein oprM | | Authors: | Akama, H, Kanemaki, M, Yoshimura, M, Tsukihara, T, Kashiwagi, T, Narita, S, Nakagawa, A, Nakae, T. | | Deposit date: | 2004-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end
J.Biol.Chem., 279, 2004
|
|
1WSV
 
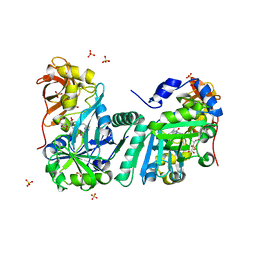 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-11 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
1X1N
 
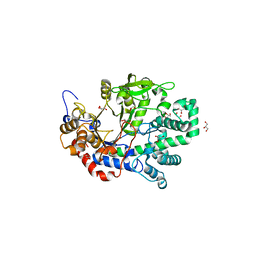 | | Structure determination and refinement at 1.8 A resolution of Disproportionating Enzyme from Potato | | Descriptor: | 4-alpha-glucanotransferase, CALCIUM ION, GLYCEROL | | Authors: | Imamura, K, Matsuura, T, Takaha, T, Fujii, K, Nakagawa, A, Kusunoki, M, Nitta, Y. | | Deposit date: | 2005-04-08 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination and refinement at 1.8 A resolution of Disproportionating Enzyme from Potato
to be published
|
|
1WSR
 
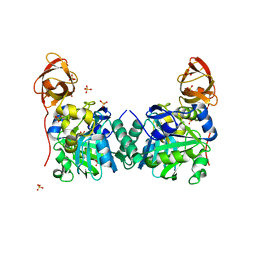 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
