6AGB
 
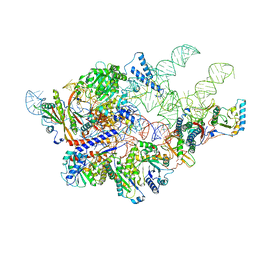 | | Cryo-EM structure of yeast Ribonuclease P | | Descriptor: | RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, Ribonuclease P protein subunit RPR2, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
6AS7
 
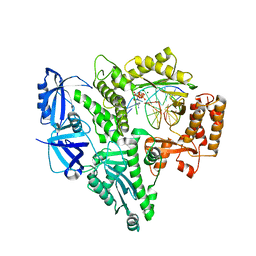 | | CRYSTAL STRUCTURE OF THE CATALYTIC CORE OF HUMAN DNA POLYMERASE ALPHA IN TERNARY COMPLEX WITH AN DNA-PRIMED DNA TEMPLATE AND DCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, COBALT (II) ION, DNA (5'-D(*AP*GP*GP*CP*GP*CP*TP*CP*CP*AP*GP*GP*C)-3'), ... | | Authors: | Tahirov, T.H, Baranovskiy, A.G, Babayeva, N.D. | | Deposit date: | 2017-08-23 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Activity and fidelity of human DNA polymerase alpha depend on primer structure.
J. Biol. Chem., 293, 2018
|
|
3I5N
 
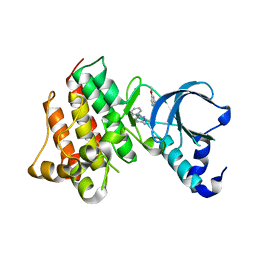 | | Crystal structure of c-Met with triazolopyridazine inhibitor 13 | | Descriptor: | 7-methoxy-N-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]-1,5-naphthyridin-4-amine, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.M, Boezio, A.A. | | Deposit date: | 2009-07-06 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of potent and selective triazolopyridazine series of c-Met inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E7X
 
 | |
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7VSI
 
 | | Structure of human SGLT2-MAP17 complex bound with empagliflozin | | Descriptor: | (2S,3R,4R,5S,6R)-2-[4-chloranyl-3-[[4-[(3S)-oxolan-3-yl]oxyphenyl]methyl]phenyl]-6-(hydroxymethyl)oxane-3,4,5-triol, PALMITIC ACID, PDZK1-interacting protein 1, ... | | Authors: | Chen, L, Niu, Y, Liu, R. | | Deposit date: | 2021-10-26 | | Release date: | 2021-12-15 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of inhibition of the human SGLT2-MAP17 glucose transporter.
Nature, 601, 2022
|
|
5YZ2
 
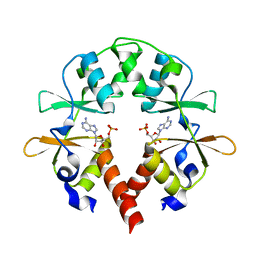 | | the cystathionine-beta-synthase (CBS) domain of magnesium and cobalt efflux protein CorC in complex with both C2'- and C3'-endo AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein CorC | | Authors: | Feng, N, Qi, C, Li, D.F, Wang, D.C. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C2'- and C3'-endo equilibrium for AMP molecules bound in the cystathionine-beta-synthase domain.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5Y36
 
 | | Cryo-EM structure of SpCas9-sgRNA-DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, complementary DNA strand, ... | | Authors: | Huang, Q, Li, G, Huai, C. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural insights into DNA cleavage activation of CRISPR-Cas9 system
Nat Commun, 8, 2017
|
|
5EJ2
 
 | |
5ZBR
 
 | | Crystal Structure of Kinesin-3 KIF13B motor domain in AMPPNP form | | Descriptor: | Kinesin family member 13B, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J.Q, Wang, S, Feng, W. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Delineation of the Neck Linker of Kinesin-3 for Processive Movement.
J. Mol. Biol., 430, 2018
|
|
2XT4
 
 | |
6KLS
 
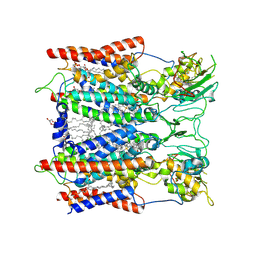 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, Cytochrome c, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | Authors: | Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
7KN4
 
 | |
7FAH
 
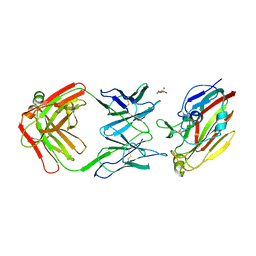 | | Immune complex of head region of CA09 HA and neutralizing antibody 12H5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Li, T.T, Xue, W.H, Gu, Y, Li, S.W. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
4CYW
 
 | | Structure of the A_mallard_Sweden_51_2002 H10 Avian Haemmaglutinin in complex with human receptor analog 6-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Martin, S.R, Haire, L.F, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-04-15 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Receptor Binding by H10 Influenza Viruses.
Nature, 511, 2014
|
|
4CYV
 
 | | Structure of the A_mallard_Sweden_51_2002 H10 Avian Haemmaglutinin | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Martin, S.R, Haire, L.F, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-04-15 | | Release date: | 2014-06-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Receptor Binding by H10 Influenza Viruses.
Nature, 511, 2014
|
|
4CYZ
 
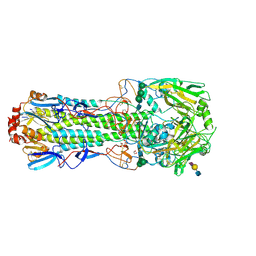 | | Structure of the A_mallard_Sweden_51_2002 H10 Avian Haemmaglutinin in complex with avian receptor analog LSTA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Martin, S.R, Haire, L.F, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Receptor Binding by H10 Influenza Viruses.
Nature, 511, 2014
|
|
2IC8
 
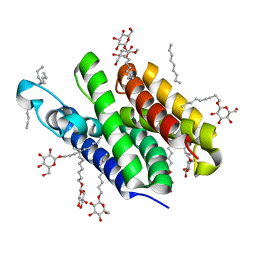 | | Crystal structure of GlpG | | Descriptor: | Protein glpG, nonyl beta-D-glucopyranoside | | Authors: | Ha, Y. | | Deposit date: | 2006-09-12 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a rhomboid family intramembrane protease.
Nature, 444, 2006
|
|
3A0E
 
 | | Crystal Structure of Polygonatum cyrtonema lectin (PCL) complexed with dimannoside | | Descriptor: | Mannose/sialic acid-binding lectin, SULFATE ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2009-03-16 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of a novel anti-HIV mannose-binding lectin from Polygonatum cyrtonema Hua with unique ligand-binding property and super-structure
J.Struct.Biol., 171, 2010
|
|
4CZ0
 
 | | Structure of the A_mallard_Sweden_51_2002 H10 Avian Haemmaglutinin in complex with avian receptor analog Su-3SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Martin, S.R, Haire, L.F, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Receptor Binding by H10 Influenza Viruses.
Nature, 511, 2014
|
|
3A0C
 
 | |
3A0D
 
 | |
8ZPS
 
 | | Cryo-EM structure of prolactin-releasing peptide recognition with Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha-1, ... | | Authors: | Zhao, L, Li, Y, Yuan, Q, Xu, H.E. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Molecular mechanism of prolactin-releasing peptide recognition and signaling via its G protein-coupled receptor.
Cell Discov, 10, 2024
|
|
