1V2R
 
 | | Trypsin inhibitor in complex with bovine trypsin variant X(SSRI)bT.B4 | | Descriptor: | CALCIUM ION, METHYL N-[(4-METHYLPHENYL)SULFONYL]GLYCYL-3-[AMINO(IMINO)METHYL]-D-PHENYLALANINATE, SULFATE ION, ... | | Authors: | Rauh, D, Klebe, G, Stubbs, M.T. | | Deposit date: | 2003-10-17 | | Release date: | 2004-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Understanding protein-ligand interactions: the price of protein flexibility
J.Mol.Biol., 335, 2004
|
|
1OXN
 
 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AEAVPWKSE peptide, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | Deposit date: | 2003-04-03 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
1OXQ
 
 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AVPIAQKSE (Smac) peptide, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | Deposit date: | 2003-04-03 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
2HFI
 
 | | Solution NMR Structure of Protein yppE from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR213 | | Descriptor: | Hypothetical protein yppE | | Authors: | Liu, G, Singarapu, K.K, Parish, D, Eletsky, A, Xu, D, Sukumaran, D, Ho, C.K, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-23 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of hypothetical protein yppE: Northeast Structural Genomics Consortium Target SR213
TO BE PUBLISHED
|
|
4PL3
 
 | | Crystal structure of murine IRE1 in complex with MKC9989 inhibitor | | Descriptor: | 7-hydroxy-6-methoxy-3-[2-(2-methoxyethoxy)ethyl]-4,8-dimethyl-2H-chromen-2-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sanches, M, Duffy, N, Talukdar, M, Thevakumaran, N, Chiovitti, D, Al-awar, R, Patterson, J.B, Sicheri, F. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors.
Nat Commun, 5, 2014
|
|
6PLG
 
 | | Crystal structure of human PHGDH complexed with Compound 15 | | Descriptor: | (2S)-(4-{3-[(4,5-dichloro-1-methyl-1H-indole-2-carbonyl)amino]oxetan-3-yl}phenyl)(pyridin-3-yl)acetic acid, D-3-phosphoglycerate dehydrogenase, D-MALATE | | Authors: | Olland, A, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2019-06-30 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Inhibition of 3-phosphoglycerate dehydrogenase (PHGDH) by indole amides abrogates de novo serine synthesis in cancer cells.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8B6U
 
 | | Mpf2Ba1 monomer | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez-Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
8B6W
 
 | | Mpf2Ba1 pore | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
8B6V
 
 | | Mp2Ba1 pre-pore | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
6QXU
 
 | | Human TNKS1 in complex with 6,8-Difluoro-2-[4-(1-hydroxy-1-methyl-ethyl)-phenyl]-3H-quinazolin-4-one | | Descriptor: | 6,8-bis(fluoranyl)-2-[4-(2-oxidanylpropan-2-yl)phenyl]-3~{H}-quinazolin-4-one, BETA-MERCAPTOETHANOL, Poly [ADP-ribose] polymerase tankyrase-1, ... | | Authors: | Musil, D, Lehmann, D, Buchstaller, H.-P. | | Deposit date: | 2019-03-08 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery and Optimization of 2-Arylquinazolin-4-ones into a Potent and Selective Tankyrase Inhibitor Modulating Wnt Pathway Activity.
J.Med.Chem., 62, 2019
|
|
6P0J
 
 | | Crystal structure of GDP-bound human RalA | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Ral-A | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8BBQ
 
 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, GLYCEROL, ... | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
8BBR
 
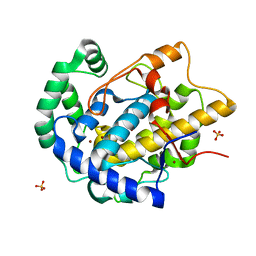 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, SULFATE ION | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
3DSD
 
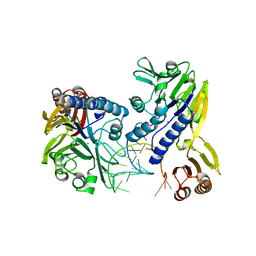 | | Crystal structure of P. furiosus Mre11-H85S bound to a branched DNA and manganese | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DGP*DAP*DTP*DA)-3'), DNA double-strand break repair protein mre11, MANGANESE (II) ION | | Authors: | Williams, R.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
6FJS
 
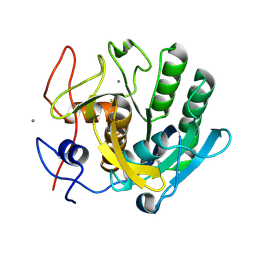 | | Proteinase~K SIRAS phased structure of room-temperature, serially collected synchrotron data | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Botha, S, Baitan, D, Jungnickel, K.E.J, Oberthuer, D, Schmidt, C, Stern, S, Wiedorn, M.O, Perbandt, M, Chapman, H.N, Betzel, C. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novoprotein structure determination by heavy-atom soaking in lipidic cubic phase and SIRAS phasing using serial synchrotron crystallography.
IUCrJ, 5, 2018
|
|
6PBW
 
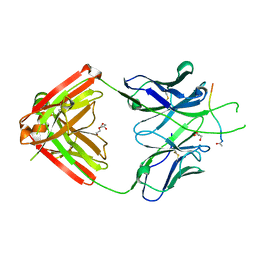 | | Crystal structure of Fab667 complex | | Descriptor: | Fab667 heavy chain, Fab667 light chain, GLYCEROL, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2019-06-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.058 Å) | | Cite: | Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein.
Plos Pathog., 16, 2020
|
|
5E79
 
 | | Macromolecular diffractive imaging using imperfect crystals | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthur, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffer, A, Dorner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
5EBZ
 
 | | Crystal structure of human IKK1 | | Descriptor: | 2,3-di-O-sulfo-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-2,4-di-O-sulfo-alpha-D-glucopyranose, 2-azanyl-5-phenyl-3-(4-sulfamoylphenyl)benzamide, Inhibitor of nuclear factor kappa-B kinase subunit alpha, ... | | Authors: | Polley, S, Passos, D, Huang, D, Biswas, T, Verma, I, Lyumkis, D, Ghosh, G. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
3DMK
 
 | | Crystal structure of Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, N-terminal eight Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, ... | | Authors: | Sawaya, M.R, Wojtowicz, W.M, Eisenberg, D, Zipursky, S.L. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms.
Cell(Cambridge,Mass.), 134, 2008
|
|
3DS6
 
 | | P38 complex with a phthalazine inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-[1-(2-methylphenyl)phthalazin-6-yl]benzamide | | Authors: | Herberich, B, Syed, R, Li, V, Grosfeld, D. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of highly selective and potent p38 inhibitors based on a phthalazine scaffold.
J.Med.Chem., 51, 2008
|
|
5DUD
 
 | | Crystal structure of E. coli YbgJK | | Descriptor: | YbgJ, YbgK | | Authors: | Arbing, M.A, Kaufmann, M, Shin, A, Medrano-Soto, A, Cascio, D, Eisenberg, D. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of E. coli YbgJK
To Be Published
|
|
5HRY
 
 | | Computationally Designed Cyclic Dimer ank3C2_1 | | Descriptor: | ank3C2_1 | | Authors: | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | Deposit date: | 2016-01-24 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5EFQ
 
 | | Crystal structure of human Cdk13/Cyclin K in complex with ADP-aluminum fluoride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin-K, ... | | Authors: | Hoenig, D, Greifenberg, A.K, Anand, K, Geyer, M. | | Deposit date: | 2015-10-24 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of the Cdk13/Cyclin K Complex.
Cell Rep, 14, 2016
|
|
6PBV
 
 | | Crystal structure of Fab668 complex | | Descriptor: | 1,2-ETHANEDIOL, Fab668 heavy chain, Fab668 light chain, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2019-06-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.566 Å) | | Cite: | Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein.
Plos Pathog., 16, 2020
|
|
6PLF
 
 | | Crystal structure of human PHGDH complexed with Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-{(1S)-1-[(5-chloro-6-{[(5S)-2-oxo-1,3-oxazolidin-5-yl]methoxy}-1H-indole-2-carbonyl)amino]-2-hydroxyethyl}benzoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Olland, A, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2019-06-30 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of 3-phosphoglycerate dehydrogenase (PHGDH) by indole amides abrogates de novo serine synthesis in cancer cells.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
