8F1K
 
 | | SigN RNA polymerase early-melted intermediate bound to full duplex DNA fragment dhsU36 (-12T) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Mueller, A.U, Chen, J, Darst, S.A. | | Deposit date: | 2022-11-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A general mechanism for transcription bubble nucleation in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9KTI
 
 | | CryoEM structure of a 2,3-hydroxycinnamic acid 1,2-dioxygenase MhpB in substrate bound form | | Descriptor: | 2,3-dihydroxyphenylpropionate/2,3-dihydroxicinnamic acid 1,2-dioxygenase, 3-[2,3-bis(oxidanyl)phenyl]propanoic acid, FE (II) ION | | Authors: | Dong, X, Jiang, W.X, Ma, L.X, Xing, Q. | | Deposit date: | 2024-12-02 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural and catalytic insights into MhpB: A dioxygenase enzyme for degrading catecholic pollutants.
J Hazard Mater, 488, 2025
|
|
8K04
 
 | | CryoEM structure of a 2,3-hydroxycinnamic acid 1,2-dioxygenase MhpB in apo form | | Descriptor: | 2,3-dihydroxyphenylpropionate/2,3-dihydroxicinnamic acid 1,2-dioxygenase | | Authors: | Jiang, W.X, Cheng, X.Q, Ma, L.X, Xing, Q. | | Deposit date: | 2023-07-07 | | Release date: | 2024-07-10 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural and catalytic insights into MhpB: A dioxygenase enzyme for degrading catecholic pollutants.
J Hazard Mater, 488, 2025
|
|
5UI8
 
 | | structure of sigmaN-holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | Crystal structure of Aquifex aeolicus sigma (N) bound to promoter DNA and the structure of sigma (N)-holoenzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8U52
 
 | | Co-crystal structure of human transthyretin conjugated to the stilbene substructure derived from reaction with the fluorogenic covalent kinetic stabilizer A2 | | Descriptor: | S-(4-fluorophenyl) 3-(dimethylamino)-5-[(E)-2-(4-hydroxy-3,5-dimethylphenyl)ethenyl]benzenecarbothioate, Transthyretin | | Authors: | Yan, N.L, Nugroho, K, Kline, G.M, Basanta, B, Lander, G.C, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2023-09-11 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The conformational landscape of human transthyretin revealed by cryo-EM.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VSU
 
 | | Cryo-EM structure of LKB1-STRADalpha-MO25alpha heterocomplex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, Isoform 3 of STE20-related kinase adapter protein alpha, ... | | Authors: | Chan, L.M, Courteau, B.J, Verba, K.A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-10 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | High-resolution single-particle imaging at 100-200 keV with the Gatan Alpine direct electron detector.
J.Struct.Biol., 216, 2024
|
|
7N6Z
 
 | | Crystal Structure of PI5P4KIIAlpha | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7N
 
 | | Crystal Structure of PI5P4KIIAlpha complex with Volasertib | | Descriptor: | N-{trans-4-[4-(cyclopropylmethyl)piperazin-1-yl]cyclohexyl}-4-{[(7R)-7-ethyl-5-methyl-8-(1-methylethyl)-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxybenzamide, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7O
 
 | | Crystal Structure of PI5P4KIIAlpha complex with Palbociclib | | Descriptor: | 6-ACETYL-8-CYCLOPENTYL-5-METHYL-2-[(5-PIPERAZIN-1-YLPYRIDIN-2-YL)AMINO]PYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7J
 
 | | Crystal Structure of PI5P4KIIAlpha complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, ... | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7M
 
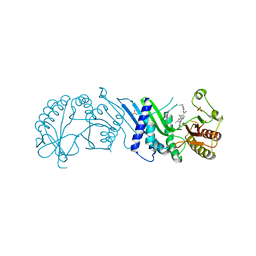 | | Crystal Structure of PI5P4KIIAlpha complex with BI-2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7K
 
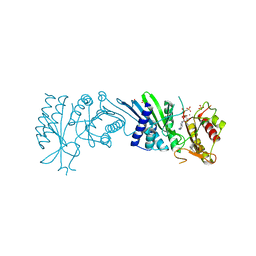 | | Crystal Structure of PI5P4KIIAlpha complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, ... | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N81
 
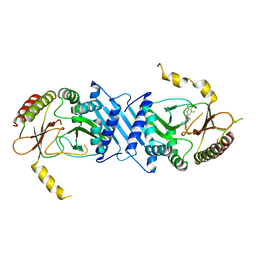 | | Crystal Structure of PI5P4KIIBeta complex with CC260 | | Descriptor: | (7R)-8-cyclopentyl-7-(cyclopentylmethyl)-2-[(3,5-dichloro-4-hydroxyphenyl)amino]-5-methyl-7,8-dihydropteridin-6(5H)-one, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-11 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7L
 
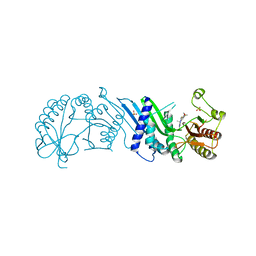 | | Crystal Structure of PI5P4KIIAlpha complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N80
 
 | | Crystal Structure of PI5P4KIIBeta | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-11 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N71
 
 | | Crystal Structure of PI5P4KIIAlpha | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7X
 
 | | Crystal structure of BCX7353(ORLADEYO) in complex with human plasma kallikrein serine protease domain at 2.1 angstrom resolution | | Descriptor: | Orladeyo, PHOSPHATE ION, Plasma kallikrein light chain | | Authors: | Krishnan, R, Yarlagadda, B.S, Kotian, P, Polach, K.J, Zhang, W. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Berotralstat (BCX7353): Structure-Guided Design of a Potent, Selective, and Oral Plasma Kallikrein Inhibitor to Prevent Attacks of Hereditary Angioedema (HAE).
J.Med.Chem., 64, 2021
|
|
7WV9
 
 | | Allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 in complex with Gi protein | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, ... | | Authors: | Xu, Z, Shao, Z. | | Deposit date: | 2022-02-10 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
5YIM
 
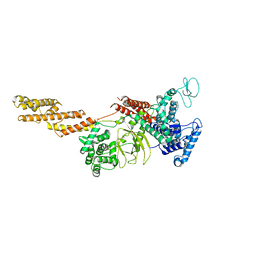 | | Structure of a Legionella effector | | Descriptor: | SdeA | | Authors: | Feng, Y, Dong, Y, Wang, W. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
5YIK
 
 | |
5YIJ
 
 | | Structure of a Legionella effector with substrates | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, SdeA, Ubiquitin | | Authors: | Feng, Y, Mu, Y, Wang, H. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
6CA7
 
 | |
6CA9
 
 | |
6CA6
 
 | |
3LX7
 
 | | Crystal structure of a Novel Tudor domain-containing protein SGF29 | | Descriptor: | SAGA-associated factor 29 homolog, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Bian, C.B, Xu, C, Tempel, W, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J. | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
