5GXB
 
 | | crystal structure of a LacY/Nanobody complex | | Descriptor: | Lactose permease, nanobody | | Authors: | Jiang, X, Wu, J.P, Yan, N, Kaback, H.R. | | Deposit date: | 2016-09-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a LacY-nanobody complex in a periplasmic-open conformation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7SP9
 
 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed channel-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SPA
 
 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed, channel-open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP6
 
 | | Chlorella virus hyaluronan synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP8
 
 | | Chlorella virus Hyaluronan Synthase bound to UDP-GlcNAc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP7
 
 | | Chlorella virus hyaluronan synthase inhibited by UDP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
5DA0
 
 | | Structure of the the SLC26 transporter SLC26Dg in complex with a nanobody | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, Nanobody, Sulphate transporter | | Authors: | Dutzler, R, Geertsma, E.R, Chang, Y, Shaik, F.R. | | Deposit date: | 2015-08-19 | | Release date: | 2015-09-09 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a prokaryotic fumarate transporter reveals the architecture of the SLC26 family.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5G5R
 
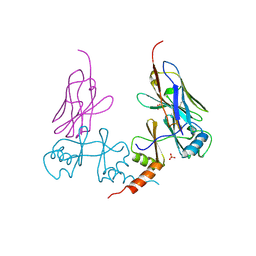 | |
5DA4
 
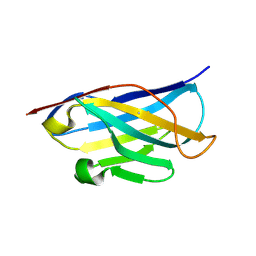 | |
5DFZ
 
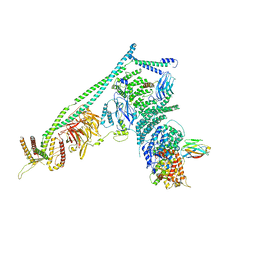 | | Structure of Vps34 complex II from S. cerevisiae. | | Descriptor: | Nanobody binding S. cerevisiae Vps34, Phosphatidylinositol 3-kinase VPS34, Putative N-terminal domain of S. cerevisiae Vps30, ... | | Authors: | Rostislavleva, K, Soler, N, Ohashi, Y, Zhang, L, Williams, R.L. | | Deposit date: | 2015-08-27 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes.
Science, 350, 2015
|
|
5G5X
 
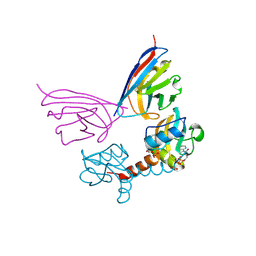 | |
5IOF
 
 | |
6HHU
 
 | |
6HER
 
 | | Mouse prion protein in complex with Nanobody 484 | | Descriptor: | GLYCEROL, Major prion protein, Nanobody 484, ... | | Authors: | Soror, S.H, Abskharon, R, Wohlkonig, A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
6HHD
 
 | | Mouse Prion Protein in complex with Nanobody 484 | | Descriptor: | CHLORIDE ION, GLYCEROL, Major prion protein, ... | | Authors: | Soror, S, Abskharon, R, Wohlkonig, A. | | Deposit date: | 2018-08-28 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
5OCL
 
 | | Nanobody-anti-VGLUT nanobody complex | | Descriptor: | anti-llama nanobody, anti-vglut nanobody | | Authors: | Dutzler, R, Schenck, S, Kunz, L. | | Deposit date: | 2017-07-03 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Generation and Characterization of Anti-VGLUT Nanobodies Acting as Inhibitors of Transport.
Biochemistry, 56, 2017
|
|
4OCN
 
 | |
5M94
 
 | |
4OCM
 
 | |
4OCL
 
 | |
7NVL
 
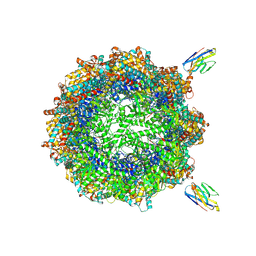 | | Human TRiC complex in closed state with nanobody bound (Consensus Map) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NVO
 
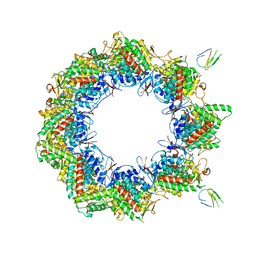 | | Human TRiC complex in open state with nanobody bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NVN
 
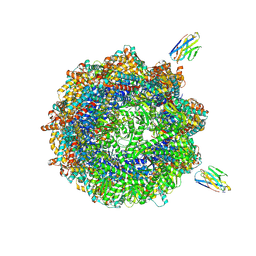 | | Human TRiC complex in closed state with nanobody and tubulin bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NVM
 
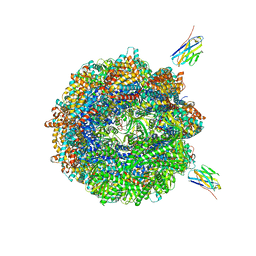 | | Human TRiC complex in closed state with nanobody Nb18, actin and PhLP2A bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Actin, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8CQB
 
 | |
