6SHX
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6SNS
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6SNV
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5HDA
 
 | |
6TQH
 
 | | Escherichia coli AdhE structure in its extended conformation | | Descriptor: | Aldehyde-alcohol dehydrogenase, FE (III) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Fronzes, R, Pony, P. | | Deposit date: | 2019-12-16 | | Release date: | 2020-06-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Filamentation of the bacterial bi-functional alcohol/aldehyde dehydrogenase AdhE is essential for substrate channeling and enzymatic regulation.
Nat Commun, 11, 2020
|
|
7BO6
 
 | | VDR complex with LCA derivative | | Descriptor: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2021-01-24 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Lithocholic acid-based design of noncalcemic vitamin D receptor agonists.
Bioorg.Chem., 111, 2021
|
|
6G1B
 
 | | Corynebacterium glutamicum OxyR, oxidized form | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, FORMIC ACID, HYDROGEN PEROXIDE, ... | | Authors: | Young, D.R, Pedre, B.P, Messens, J.M. | | Deposit date: | 2018-03-21 | | Release date: | 2018-12-05 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G4R
 
 | | Corynebacterium glutamicum OxyR C206S mutant, H2O2-bound | | Descriptor: | 1,2-ETHANEDIOL, HYDROGEN PEROXIDE, Hydrogen peroxide-inducible genes activator, ... | | Authors: | Young, D.R, Pedre, B.P, Messens, J.M. | | Deposit date: | 2018-03-28 | | Release date: | 2018-12-05 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G1D
 
 | | Corynebacterium glutamicum OxyR C206 mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Young, D.R, Pedre, B.P, Messens, J.M. | | Deposit date: | 2018-03-21 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ZCE
 
 | |
7ZCF
 
 | |
6E99
 
 | |
6ED6
 
 | |
6E9W
 
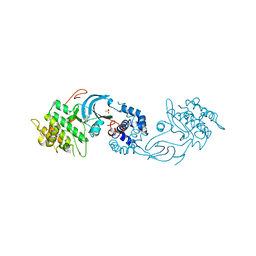 | | Crystal structure of Rock1 with a pyridinylbenzamide based inhibitor | | Descriptor: | N-[(2,3-dihydro-1,4-benzodioxin-5-yl)methyl]-4-(pyridin-4-yl)benzamide, Rho-associated protein kinase 1, SULFATE ION | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification of Selective Dual ROCK1 and ROCK2 Inhibitors Using Structure Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6E9L
 
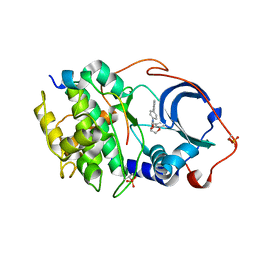 | |
8AMD
 
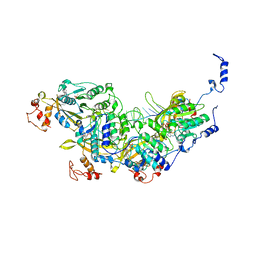 | | Cryo-EM structure of the RecA presynaptic filament from S.pneumoniae | | Descriptor: | DNA, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein RecA | | Authors: | Perry, T.N, Fronzes, R, Polard, P, Hertzog, M. | | Deposit date: | 2022-08-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Assembly mechanism and cryoEM structure of RecA recombination nucleofilaments from Streptococcus pneumoniae.
Nucleic Acids Res., 51, 2023
|
|
8AMF
 
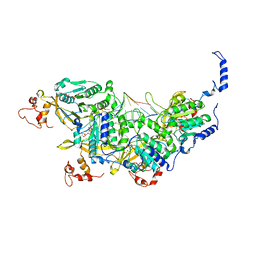 | | Cryo-EM structure of the RecA postsynaptic filament from S. pneumoniae | | Descriptor: | DNA, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein RecA | | Authors: | Perry, T.N, Fronzes, R, Polard, P, Hertzog, M. | | Deposit date: | 2022-08-03 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Assembly mechanism and cryoEM structure of RecA recombination nucleofilaments from Streptococcus pneumoniae.
Nucleic Acids Res., 51, 2023
|
|
8TX6
 
 | |
8TX5
 
 | |
6BW3
 
 | | Crystal structure of RBBP4 in complex with PRDM3 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, MDS1 and EVI1 complex locus protein MDS1, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
6BW4
 
 | | Crystal structure of RBBP4 in complex with PRDM16 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, PR domain zinc finger protein 16, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
7QGC
 
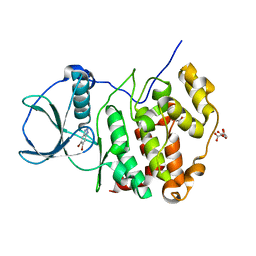 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 5.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, CITRATE ANION, Casein kinase II subunit alpha, ... | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
7QGE
 
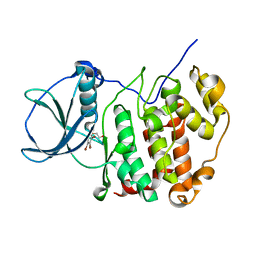 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6,7,8-TETRABROMOBENZOTRIAZOLE (TBBt) AT PH 8.5 | | Descriptor: | 4,5,6,7-TETRABROMOBENZOTRIAZOLE, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
7QGD
 
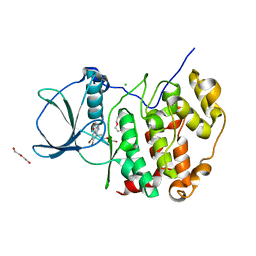 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 8.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
7QGB
 
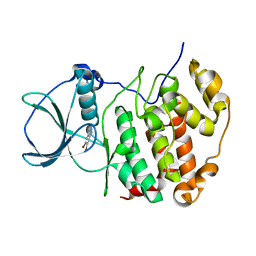 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 6.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, Casein kinase II subunit alpha | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
