6X22
 
 | |
5UKQ
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522.2 Fab | | Descriptor: | DH522.2 Fab fragment heavy chain, DH522.2 Fab fragment light chain, GLYCEROL | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
2L6C
 
 | |
4UP7
 
 | |
4UPA
 
 | |
6X1N
 
 | |
4UP8
 
 | |
6X20
 
 | |
4UP9
 
 | |
3KRR
 
 | | Crystal Structure of JAK2 complexed with a potent quinoxaline ATP site inhibitor | | Descriptor: | 8-[3,5-difluoro-4-(morpholin-4-ylmethyl)phenyl]-2-(1-piperidin-4-yl-1H-pyrazol-4-yl)quinoxaline, Tyrosine-protein kinase JAK2 | | Authors: | Tavares, G.A, Gerspacher, M, Kroemer, M, Scheufler, C. | | Deposit date: | 2009-11-19 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and Selective Inhibition of Polycythemia by the Quinoxaline JAK2 Inhibitor NVP-BSK805
Mol.Cancer Ther., 9, 2010
|
|
2L6D
 
 | |
6TYL
 
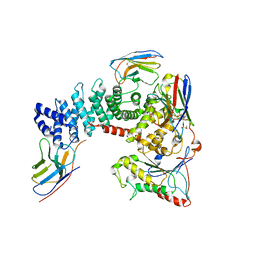 | | Crystal structure of mammalian Ric-8A:Galpha(i):nanobody complex | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, Nanobody A, Nanobody B, ... | | Authors: | Mou, T.C, McClelland, L, Yates-Hansen, C, Sprang, S.R. | | Deposit date: | 2019-08-09 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to G alpha i1.
Nat Commun, 11, 2020
|
|
7Q55
 
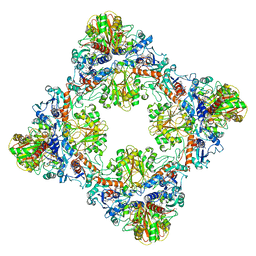 | | Single Particle Cryo-EM structure of photosynthetic A8B8 glyceraldehyde-3-phosphate dehydrogenase hexadecamer (major conformer) from Spinacia oleracia. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q56
 
 | | Single Particle Cryo-EM structure of photosynthetic A8B8 glyceraldehyde-3-phosphate dehydrogenase (minor conformer) from Spinacia oleracea. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q57
 
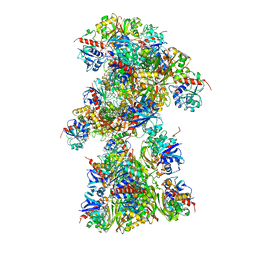 | | Single Particle Cryo-EM structure of photosynthetic A10B10 glyceraldehyde-3-phospahte dehydrogenase from Spinacia oleracea. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q53
 
 | | Single Particle Cryo-EM structure of photosynthetic A2B2 glyceraldehyde 3-phosphate dehydrogenase from Spinacia oleracia | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZTY
 
 | |
7QUO
 
 | | FimH lectin domain in complex with oligomannose-6 | | Descriptor: | FimH, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Bouckaert, J, Bourenkov, G.P. | | Deposit date: | 2022-01-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors
J.Biol.Chem., 299, 2023
|
|
7RD1
 
 | | The Capsid Structure of the ChAdOx1 viral vector/chimpanzee adenovirus Y25 | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Baker, A.T, Boyd, R.J, Sarkar, D, Vermaas, J.V, Williams, D, Singharoy, A. | | Deposit date: | 2021-07-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome.
Sci Adv, 7, 2021
|
|
7Q54
 
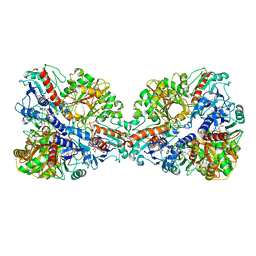 | | Single Particle Cryo-EM structure of photosynthetic A4B4-glyceraldehyde 3-phosphate dehydrogenase from Spinacia oleracia. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
2K8X
 
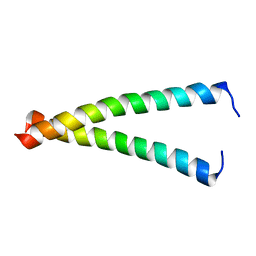 | |
5U39
 
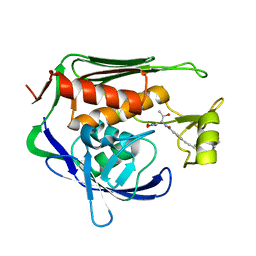 | | Pseudomonas aeruginosa LpxC in complex with CHIR-090 | | Descriptor: | N-{(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl]propyl}-4-{[4-(morpholin-4-ylmethyl)phenyl]ethynyl}benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Sprague, E.R. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC.
J. Med. Chem., 60, 2017
|
|
7A5X
 
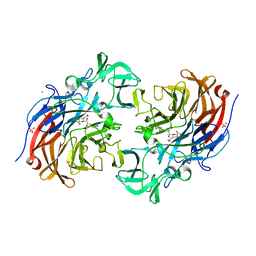 | | Two copies of the catalytic domain of NanA sialidase from Streptococcus pneumoniae juxtaposed in the P212121 space group, in complex with DANA derivatized with a PEG linker on the glycerol group. | | Descriptor: | (4~{S},5~{R},6~{R})-5-acetamido-6-[(1~{R},2~{S})-3-[[1-(2-ethoxyethyl)-1,2,3-triazol-4-yl]methylsulfanyl]-1,2-bis(oxidanyl)propyl]-4-oxidanyl-oxane-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bridot, C, Bouckaert, J. | | Deposit date: | 2020-08-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Polyvalent Transition-State Analogues of Sialyl Substrates Strongly Inhibit Bacterial Sialidases*.
Chemistry, 27, 2021
|
|
7BHD
 
 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2021-01-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
5U3B
 
 | | Pseudomonas aeruginosa LpxC in complex with NVS-LPXC-01 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-[(2S)-3-amino-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-[(but-2-yn-1-yl)oxy]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Sprague, E.R. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC.
J. Med. Chem., 60, 2017
|
|
