1MSZ
 
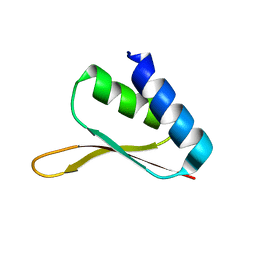 | | Solution structure of the R3H domain from human Smubp-2 | | Descriptor: | DNA-binding protein SMUBP-2 | | Authors: | Liepinsh, E, Leonchiks, A, Sharipo, A, Guignard, L, Otting, G. | | Deposit date: | 2002-09-20 | | Release date: | 2002-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the R3H domain from human Smubp-2
J.Mol.Biol., 326, 2003
|
|
1JBI
 
 | | NMR structure of the LCCL domain | | Descriptor: | cochlin | | Authors: | Liepinsh, E, Trexler, M, Kaikkonen, A, Weigelt, J, Banyai, L, Patthy, L, Otting, G. | | Deposit date: | 2001-06-05 | | Release date: | 2001-10-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the LCCL domain and implications for DFNA9 deafness disorder.
EMBO J., 20, 2001
|
|
2HAJ
 
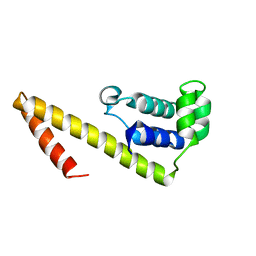 | |
2K7R
 
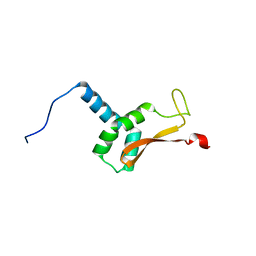 | | N-terminal domain of the Bacillus subtilis helicase-loading protein DnaI | | Descriptor: | Primosomal protein dnaI, ZINC ION | | Authors: | Loscha, K.V, Jaudzems, K, Ioannou, C, Su, X.C, Hill, F.R, Otting, G, Dixon, N.E, Liepinsh, E. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Nucleic Acids Res., 37, 2009
|
|
4NYH
 
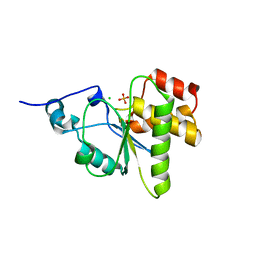 | | Orthorhombic crystal form of pir1 dual specificity phosphatase core | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, RNA/RNP complex-1-interacting phosphatase | | Authors: | Sankhala, R.S, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2013-12-10 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Human PIR1, an Atypical Dual-Specificity Phosphatase.
Biochemistry, 53, 2014
|
|
2P8Q
 
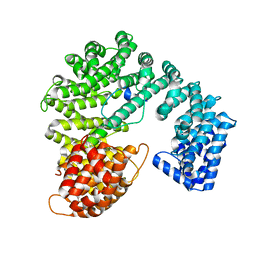 | |
2Q5D
 
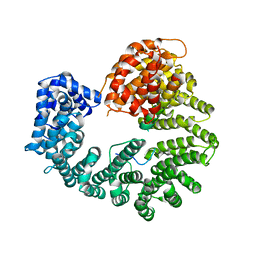 | |
2LRR
 
 | | Solution structure of the R3H domain from human Smubp-2 in complex with 2'-deoxyguanosine-5'-monophosphate | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA-binding protein SMUBP-2 | | Authors: | Jaudzems, K, Zhulenkovs, D, Otting, G, Liepinsh, E. | | Deposit date: | 2012-04-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for 5'-End-Specific Recognition of Single-Stranded DNA by the R3H Domain from Human Smubp-2
J.Mol.Biol., 12, 2012
|
|
2POH
 
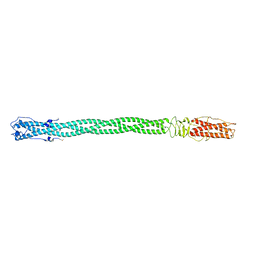 | |
2M66
 
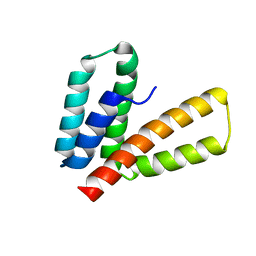 | | Endoplasmic reticulum protein 29 (ERp29) C-terminal domain: 3D Protein Fold Determination from Backbone Amide Pseudocontact Shifts Generated by Lanthanide Tags at Multiple Sites | | Descriptor: | Endoplasmic reticulum resident protein 29 | | Authors: | Yagi, H, Pilla, K, Maleckis, A, Graham, B, Huber, T, Otting, G. | | Deposit date: | 2013-03-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional protein fold determination from backbone amide pseudocontact shifts generated by lanthanide tags at multiple sites
Structure, 21, 2013
|
|
3FDS
 
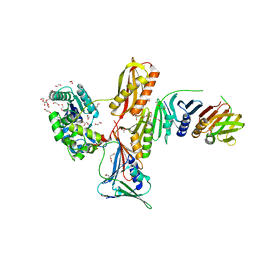 | |
4I1K
 
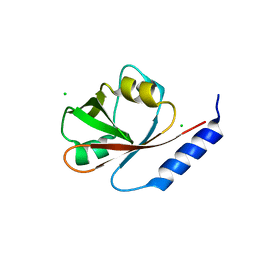 | | Crystal Structure of VRN1 (Residues 208-341) | | Descriptor: | B3 domain-containing transcription factor VRN1, CHLORIDE ION | | Authors: | King, G, Chanson, A.H, McCallum, E.J, Ohme-Takagi, M, Byriel, K, Hill, J.M, Martin, J.L, Mylne, J.S. | | Deposit date: | 2012-11-21 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Arabidopsis B3 Domain Protein VERNALIZATION1 (VRN1) Is Involved in Processes Essential for Development, with Structural and Mutational Studies Revealing Its DNA-binding Surface.
J.Biol.Chem., 288, 2013
|
|
4W7P
 
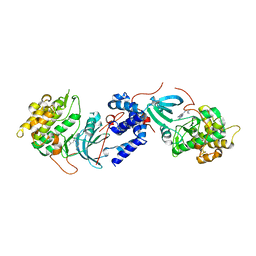 | | Crystal Structure of ROCK 1 bound to YB-15-QD37 | | Descriptor: | N~1~-[2-(1H-indazol-5-yl)pyrido[3,4-d]pyrimidin-4-yl]-2-methylpropane-1,2-diamine, Rho-associated protein kinase 1 | | Authors: | Sprague, E.R. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel ROCK inhibitors for the treatment of pulmonary arterial hypertension.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5WBP
 
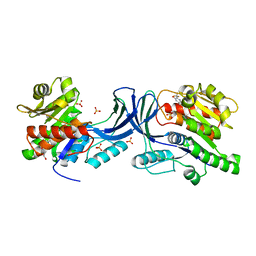 | |
5WBM
 
 | |
5WBR
 
 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-[(3S)-3-(hydroxymethyl)piperidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, GLYCEROL, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
5WBQ
 
 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 2-ethyl-7-[(3S)-3-hydroxy-3-methylpyrrolidin-1-yl]-5-(trifluoromethyl)-1H-pyrrolo[3,2-b]pyridine-6-carbonitrile, CHLORIDE ION, Ketohexokinase, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
5LNL
 
 | | Crystal structure of Hsf 1608-1749 putative domain 1 | | Descriptor: | Hsf | | Authors: | Thomsen, M, Wright, J, Ridley, J, Goldman, A. | | Deposit date: | 2016-08-05 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The crystal structure of PD1, a Haemophilus surface fibril domain.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
8D0Z
 
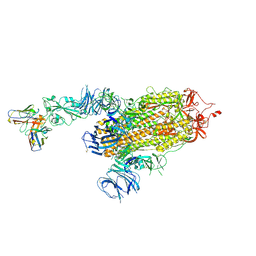 | | S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S728-1157 Fab heavy chain variable region, S728-1157 Fab light chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2022-05-26 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Site of vulnerability on SARS-CoV-2 spike induces broadly protective antibody against antigenically distinct Omicron subvariants.
J.Clin.Invest., 133, 2023
|
|
6OKD
 
 | | Crystal Structure of human transferrin receptor in complex with a cystine-dense peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Finton, K.A.K, Rupert, P.B, Strong, R.K. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A TfR-Binding Cystine-Dense Peptide Promotes Blood-Brain Barrier Penetration of Bioactive Molecules.
J.Mol.Biol., 432, 2020
|
|
9EQG
 
 | |
4DOI
 
 | |
4DOL
 
 | |
3KYM
 
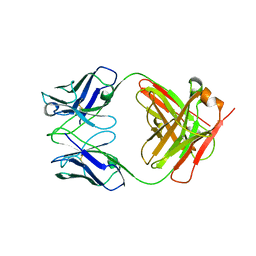 | | Crystal structure of Li33 IgG2 di-Fab | | Descriptor: | Heavy Chain Li33 IgG2, Light Chain Li33 IgG2 | | Authors: | Silvian, L.F, Pepinsky, R.B, Walus, L. | | Deposit date: | 2009-12-06 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Improving the solubility of anti-LINGO-1 monoclonal antibody Li33 by isotype switching and targeted mutagenesis.
Protein Sci., 19, 2010
|
|
3KYK
 
 | | Crystal structure of li33 Igg1 Fab | | Descriptor: | Heavy Chain Li33 IgG1, Light Chain Li33 IgG1, SULFATE ION | | Authors: | Silvian, L.F, Pepinsky, R.B, Walus, L. | | Deposit date: | 2009-12-06 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Improving the solubility of anti-LINGO-1 monoclonal antibody Li33 by isotype switching and targeted mutagenesis.
Protein Sci., 19, 2010
|
|
