7WUI
 
 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Guo, S.C, He, Q.T, Xiao, P, Sun, J.P, Yu, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
7WUQ
 
 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Q.T, Guo, S.C, Xiao, P, Sun, J.P, Yu, X, Gou, L, Kong, L.L, Zhang, L. | | Deposit date: | 2022-02-09 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
7WUJ
 
 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G4,Uncharacterized protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Guo, S.C, Huang, S.M, He, Q.T, Xiao, P, Sun, J.P, Yu, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
5W9S
 
 | | Zinc finger of human CXXC5 in complex with CpG DNA | | Descriptor: | CXXC-type zinc finger protein 5, CpG DNA fragment, SULFATE ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
8K2D
 
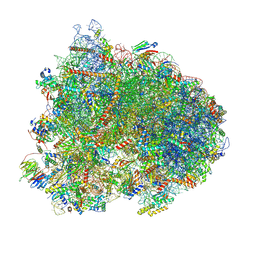 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, eEF2, Stm1 and eIF5A | | Descriptor: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Beckmann, R, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
3EPZ
 
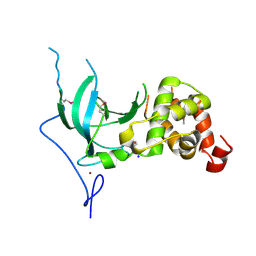 | | Structure of the replication foci-targeting sequence of human DNA cytosine methyltransferase DNMT1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, GLYCEROL, SODIUM ION, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1.
J.Biol.Chem., 286, 2011
|
|
7Y3A
 
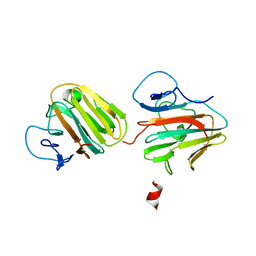 | | Crystal structure of TRIM7 bound to 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-2C | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3B
 
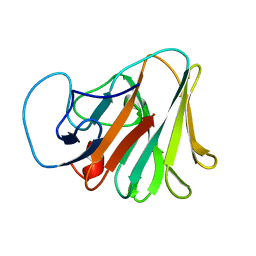 | | Crystal structure of TRIM7 bound to GN1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,TRIM7-GN1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3C
 
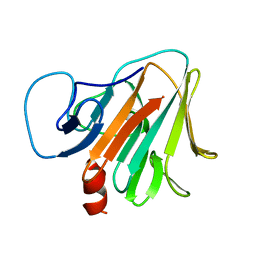 | | Crystal structure of TRIM7 bound to RACO-1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-RACO-1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2PYE
 
 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC RevealNative Diagonal Binding Geometry TCR Clone C5C1 Complexed with MHC | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Beta-2-microglobulin, Cancer/testis antigen 1B, ... | | Authors: | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | Deposit date: | 2007-05-16 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
7ESV
 
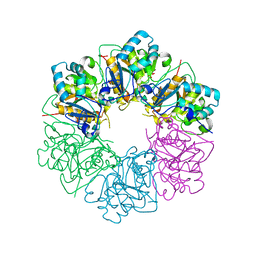 | |
7ESQ
 
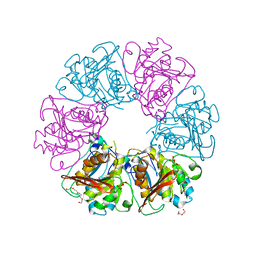 | |
7ESP
 
 | |
7ESO
 
 | |
7CE8
 
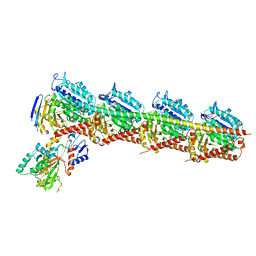 | | Crystal structure of T2R-TTL-Compound11 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
5ZZ8
 
 | | Structure of the Herpes simplex virus type 2 C-capsid with capsid-vertex-specific component | | Descriptor: | Major capsid protein, UL17, UL25, ... | | Authors: | Wang, J.L, Yuan, S, Zhu, D.J, Tang, H, Wang, N, Chen, W.Y, Gao, Q, Li, Y.H, Wang, J.Z, Liu, H.R, Zhang, X.Z, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of the herpes simplex virus type 2 C-capsid with capsid-vertex-specific component.
Nat Commun, 9, 2018
|
|
6LID
 
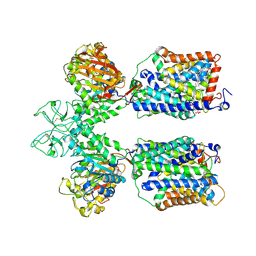 | | Heteromeric amino acid transporter b0,+AT-rBAT complex | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
6LI9
 
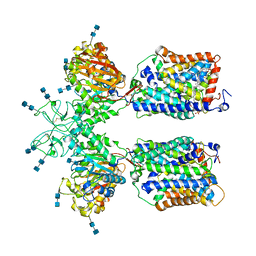 | | Heteromeric amino acid transporter b0,+AT-rBAT complex bound with Arginine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
5Z0Q
 
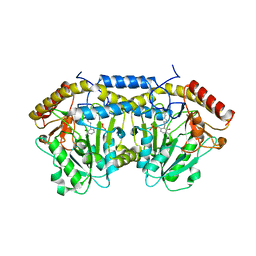 | | Crystal Structure of OvoB | | Descriptor: | Aminotransferase, class I and II, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cai, Y.J, Huang, P, Wu, L, Zhou, J.H, Liu, P.H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | In Vitro Reconstitution of the Remaining Steps in Ovothiol A Biosynthesis: C-S Lyase and Methyltransferase Reactions.
Org. Lett., 20, 2018
|
|
1K94
 
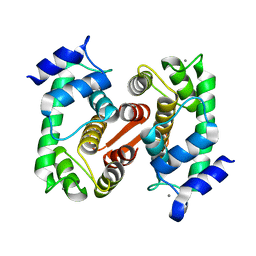 | | Crystal structure of des(1-52)grancalcin with bound calcium | | Descriptor: | CALCIUM ION, GRANCALCIN | | Authors: | Jia, J, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2001-10-26 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Ca(2+)-loaded human grancalcin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1K95
 
 | | Crystal structure of des(1-52)grancalcin with bound calcium | | Descriptor: | GRANCALCIN | | Authors: | Jia, J, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2001-10-26 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Ca(2+)-loaded human grancalcin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7K9X
 
 | |
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | Descriptor: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | Authors: | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | Deposit date: | 2021-02-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
5YF4
 
 | | A kinase complex MST4-MOB4 | | Descriptor: | MOB-like protein phocein, Peptide from Serine/threonine-protein kinase 26, ZINC ION | | Authors: | Chen, M, Zhou, Z.C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer.
J. Biol. Chem., 293, 2018
|
|
7D0C
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-3A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3A1, ... | | Authors: | Yan, R.H, Wang, R.K, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
