9D6C
 
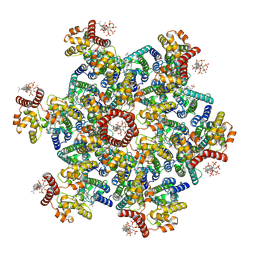 | | Gag CA-SP1 immature lattice bound with Lenacapavir and Bevirimat from enveloped virus like particles | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
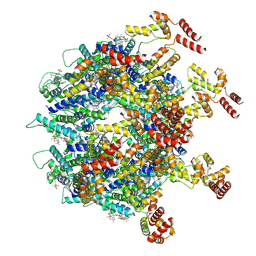
9DWD
 
 | |
6Q8Z
 
 | | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide
To Be Published
|
|
6Q91
 
 | | Structure of human galactokinase 1 bound with 5-Chloro-N-isobutyl-2-methoxybenzamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, 5-chloranyl-2-methoxy-~{N}-(2-methylpropyl)benzamide, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-17 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 5-Chloro-N-isobutyl-2-methoxybenzamide
To Be Published
|
|
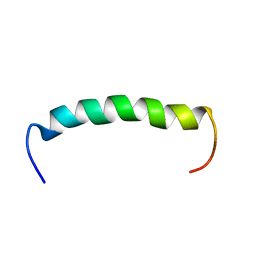
7AT7
 
 | |
5KZQ
 
 | | Metabotropic Glutamate Receptor in complex with antagonist (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
7ATB
 
 | |
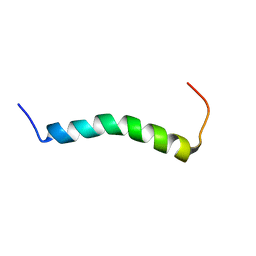
7ASY
 
 | |
1AE8
 
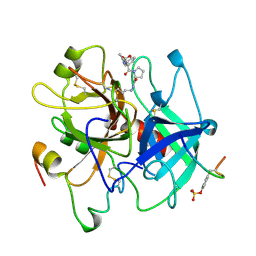 | | HUMAN ALPHA-THROMBIN INHIBITION BY EOC-D-PHE-PRO-AZALYS-ONP | | Descriptor: | 1-ETHOXYCARBONYL-D-PHE-PRO-2(4-AMINOBUTYL)HYDRAZINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
6Q3W
 
 | | Structure of human galactokinase 1 bound with Ethyl 1-(2-pyrazinyl)-4-piperidinecarboxylate | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Structure of human galactokinase 1 bound with Ethyl 1-(2-pyrazinyl)-4-piperidinecarboxylate
To Be Published
|
|
6Y3X
 
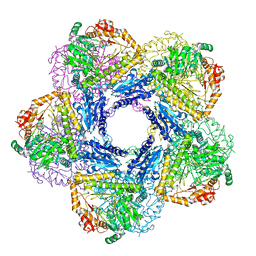 | |
7AQ1
 
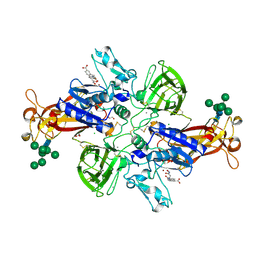 | | Crystal structure of human mature meprin beta in complex with the specific inhibitor MWT-S-270 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Linnert, M, Parthier, C, Fritz, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structure and Dynamics of Meprin beta in Complex with a Hydroxamate-Based Inhibitor.
Int J Mol Sci, 22, 2021
|
|
6ONW
 
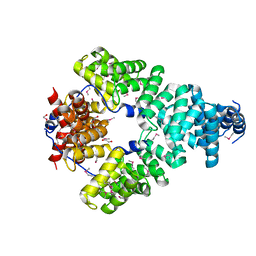 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
9I69
 
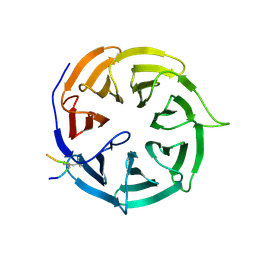 | | Crystal structure of human Cdc20 bound to synthetic D-box peptide D20 | | Descriptor: | ARG-ALA-PRO-0JY-SER-ASP, Cell division cycle protein 20 homolog | | Authors: | Eapen, R, Okoye, C, Stubbs, C, Schimpl, M, Tischer, T, Fisher, E.J, Zacharopoulou, M, Ferrer, F, Barford, D, Spring, D, Lindon, C, Phillips, C, Itzhaki, L.S. | | Deposit date: | 2025-01-29 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.463 Å) | | Cite: | Development of D-box peptides to inhibit the Anaphase Promoting Complex/Cyclosome
To be published
|
|
6ORC
 
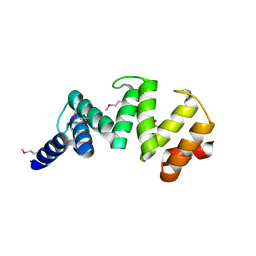 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
9I6A
 
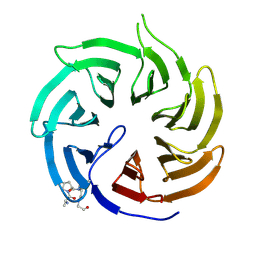 | | Crystal structure of human Cdc20 bound to synthetic D-box peptide D7 | | Descriptor: | ALA-PRO-0JY-GLY, Cell division cycle protein 20 homolog | | Authors: | Eapen, R, Okoye, C, Stubbs, C, Schimpl, M, Tischer, T, Fisher, E.J, Zacharopoulou, M, Ferrer, F, Barford, D, Spring, D, Lindon, C, Phillips, C, Itzhaki, L.S. | | Deposit date: | 2025-01-29 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Development of D-box peptides to inhibit the Anaphase Promoting Complex/Cyclosome
To be published
|
|
9I68
 
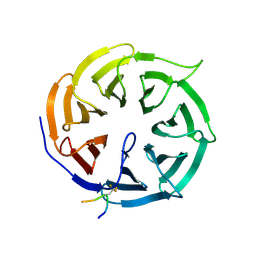 | | Crystal structure of human Cdc20 bound to synthetic D-box peptide D21 | | Descriptor: | ARG-ALA-PRO-3EG-SER-ASP, Cell division cycle protein 20 homolog | | Authors: | Eapen, R, Okoye, C, Stubbs, C, Schimpl, M, Tischer, T, Fisher, E.J, Zacharopoulou, M, Ferrer, F, Barford, D, Spring, D, Lindon, C, Phillips, C, Itzhaki, L.S. | | Deposit date: | 2025-01-29 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Development of D-box peptides to inhibit the Anaphase Promoting Complex/Cyclosome
To be published
|
|
4Y7B
 
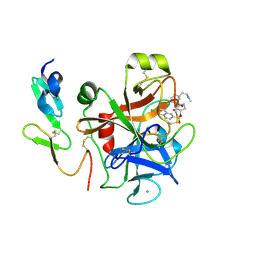 | | Factor Xa complex with GTC000441 | | Descriptor: | 6-chloro-N-{(3S)-1-[(2S)-1-{(1R,5S)-7-[2-(methylamino)ethyl]-3,7-diazabicyclo[3.3.1]non-3-yl}-1-oxopropan-2-yl]-2-oxopy rrolidin-3-yl}naphthalene-2-sulfonamide, CALCIUM ION, Coagulation factor X | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Factor Xa complex with GTC000441
to be published
|
|
6QE6
 
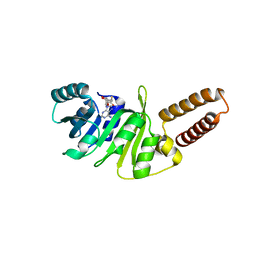 | | Structure of M. capricolum TrmK in complex with the natural cofactor product S-adenosyl-homocysteine (SAH) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
7B62
 
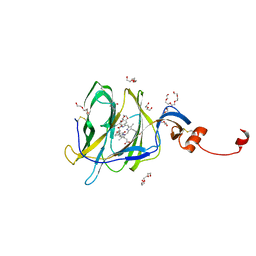 | | Crystal structure of SARS-CoV-2 spike protein N-terminal domain in complex with biliverdin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pye, V.E, Rosa, A, Roustan, C, Cherepanov, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
6VQ5
 
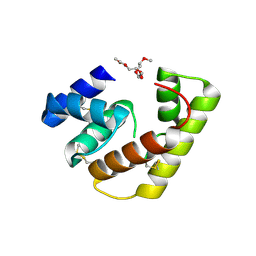 | |
5LWP
 
 | | Discovery of phenoxyindazoles and phenylthioindazoles as RORg inverse agonists | | Descriptor: | 4-[3-[2-chloranyl-6-(trifluoromethyl)phenoxy]-5-(dimethylcarbamoyl)indazol-1-yl]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Ouvry, G, Bouix-Peter, C, Ciesielski, F, Chantalat, L, Christin, O, Comino, C, Duvert, D, Feret, C, Harris, C.S, Luzy, A.-P, Musicki, B, Orfila, D, Pascau, J, Parnet, V, Perrin, A, Pierre, R, Raffin, C, Rival, Y, Taquet, N, Thoreau, E, Hennequin, L.F. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of phenoxyindazoles and phenylthioindazoles as ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8D8O
 
 | | Cryo-EM structure of substrate unbound PAPP-A | | Descriptor: | Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
6OK3
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6QE5
 
 | | Structure of E.coli RlmJ in complex with the natural cofactor product S-adenosyl-homocysteine (SAH) | | Descriptor: | Ribosomal RNA large subunit methyltransferase J, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
