8DCS
 
 | | Cryo-EM structure of cyanopindolol-bound beta1-adrenergic receptor in complex with heterotrimeric Gs-protein | | Descriptor: | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, Endolysin,Endolysin,Beta-1 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Su, M, Paknejad, N, Hite, R.K, Huang, X.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of beta 1 -adrenergic receptor in complex with Gs and ligands of different efficacies.
Nat Commun, 13, 2022
|
|
8DCR
 
 | | Cryo-EM structure of dobutamine-bound beta1-adrenergic receptor in complex with heterotrimeric Gs-protein | | Descriptor: | DOBUTAMINE, Endolysin,Endolysin,Beta-1 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Su, M, Paknejad, N, Hite, R.K, Huang, X.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of beta 1 -adrenergic receptor in complex with Gs and ligands of different efficacies.
Nat Commun, 13, 2022
|
|
3ZXH
 
 | |
6OVC
 
 | | hMcl1 inhibitor complex | | Descriptor: | (2S)-N-(benzylsulfonyl)-4-(cyclobutylmethyl)-2-(2,4-dichlorophenyl)-3,4-dihydro-2H-1,4-benzoxazine-6-carboxamide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Poppe, L. | | Deposit date: | 2019-05-07 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
7C83
 
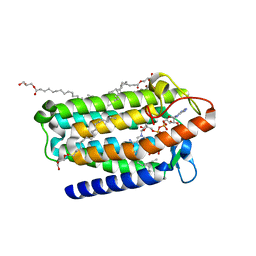 | | Crystal structure of an integral membrane steroid 5-alpha-reductase PbSRD5A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ren, R.B, Han, Y.F, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-05-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of steroid reductase SRD5A reveals conserved steroid reduction mechanism.
Nat Commun, 12, 2021
|
|
5FCS
 
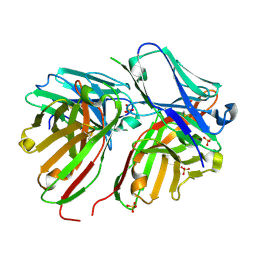 | | Diabody | | Descriptor: | Diabody, SULFATE ION | | Authors: | Mosyak, L, Root, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Development of PF-06671008, a Highly Potent Anti-P-cadherin/Anti-CD3 Bispecific DART Molecule with Extended Half-Life for the Treatment of Cancer.
Antibodies, 5, 2016
|
|
6OQN
 
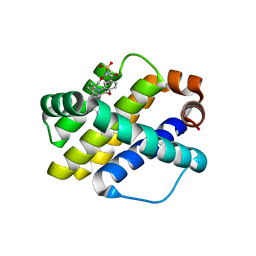 | | Crystal structure of Mcl1 with inhibitor 7 | | Descriptor: | (4S)-5'-chloro-2',3',7,8,9,10,11,12-octahydro-3H,5H,14H-spiro[1,19-etheno-16lambda~6~-[1,4]oxazepino[3,4-i][1,4,5,10]oxathiadiazacyclohexadecine-4,1'-indene]-16,16,18(15H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
6OQB
 
 | | Co-crystal structure of Mcl1 with inhibitor 10 | | Descriptor: | (4S,7aR,9aR,10S,11E,15R)-6'-chloro-15-ethyl-10-hydroxy-3',4',7a,8,9,9a,10,13,14,15-decahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)-16lambda~6~-cyclobuta[i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-4,1'-naphthalene]-16,16,18(7H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
6O6G
 
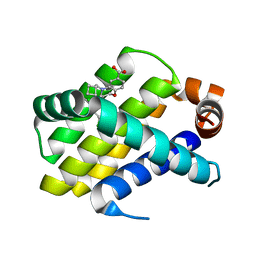 | | Co-crystal structure of Mcl1 with inhibitor | | Descriptor: | (3S)-5-(cyclobutylmethyl)-3-(2,4-dichlorophenyl)-2,3,4,5-tetrahydro-1,5-benzoxazepine-7-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-03-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
6OQD
 
 | | Crystal structure of Mcl1 with inhibitor 8 | | Descriptor: | (4S,7aR,9aR,10S,15R)-6'-chloro-10-hydroxy-15-methyl-3',4',7a,8,9,9a,10,11,12,13,14,15-dodecahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)-16lambda~6~-cyclobuta[i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-4,1'-naphthalene]-16,16,18(7H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
8K2T
 
 | |
8KBX
 
 | | Cryo-EM structure of human ATG2A-WIPI4 complex | | Descriptor: | Autophagy-related protein 2 homolog A, WD repeat domain phosphoinositide-interacting protein 4 | | Authors: | Wang, Y, Stjepanovic, G. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for lipid transfer by the ATG2A-ATG9A complex.
Nat.Struct.Mol.Biol., 2024
|
|
8KBZ
 
 | |
8KBY
 
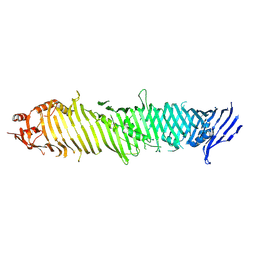 | | Cryo-EM structure of ATG2A | | Descriptor: | Autophagy-related protein 2 homolog A | | Authors: | Wang, Y, Stjepanovic, G. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for lipid transfer by the ATG2A-ATG9A complex.
Nat.Struct.Mol.Biol., 2024
|
|
8KC3
 
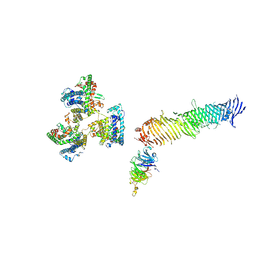 | |
8GSD
 
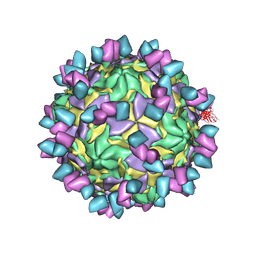 | | Echovirus3 full particle in complex with 6D10 Fab | | Descriptor: | Genome polyprotein, Genome polyprotein (Fragment), Heavy chain of 6D10, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSF
 
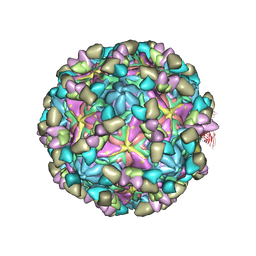 | | Echovirus3 empty particle in complex with 6D10 Fab (sideling) | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP0, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSC
 
 | | Echovirus3 A-particle in complex with 6D10 Fab | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP1, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSE
 
 | | Echovirus3 capsid protein in complex with 6D10 Fab (upright) | | Descriptor: | Heavy chain of 6D10, Light chain of 6D10, VP0, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8THS
 
 | |
6M2Z
 
 | | Crystal structure of a formolase, BFD variant M3 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
6M2Y
 
 | | Crystal structure of a formolase, BFD variant M6 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
7CAI
 
 | | SARS-CoV-2 S trimer with two RBDs in the open state and complexed with two H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
8J2Y
 
 | | Acidimicrobiaceae bacterium photocobilins protein, dark state | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
8J2X
 
 | | Saccharothrix syringae photocobilins protein, light state | | Descriptor: | BILIVERDINE IX ALPHA, COBALAMIN, Cobalamin-binding protein, ... | | Authors: | Zhang, S, Poddar, H, Levy, C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
