1TU5
 
 | | Crystal structure of bovine plasma copper-containing amine oxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lunelli, M, Di Paolo, M.L, Biadene, M, Calderone, V, Scarpa, M, Battistutta, R, Rigo, A, Zanotti, G. | | Deposit date: | 2004-06-24 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of amine oxidase from bovine serum.
J.Mol.Biol., 346, 2005
|
|
6OLU
 
 | | RIAM RA-PH core structure in the P212121 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-04-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphorylation of RIAM by src promotes integrin activation by unmasking the PH domain of RIAM.
Structure, 29, 2021
|
|
6O6H
 
 | | RIAM cc-RA-PH structure in the P21212 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphorylation of RIAM by Src Promotes Integrin Activation by Unmasking the PH Domain of RIAM.
Structure, 2020
|
|
5VCL
 
 | | Structure of the Qdm peptide bound to Qa-1a | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H2-T23 protein, ... | | Authors: | Ying, G, Zajonc, D.M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Qa-1a with bound Qa-1 determinant modifier peptide.
PLoS ONE, 12, 2017
|
|
7LLB
 
 | | Crystal structure of KPC-2 S70G/T215P mutant with hydrolyzed meropenem | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
7LLH
 
 | | KPC-2 F72Y mutant with acylated imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
7LJK
 
 | | Crystal structure of the deacylation deficient KPC-2 F72Y mutant | | Descriptor: | Beta-lactamase | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
7C8E
 
 | | Crystal Structure of 14-3-3 epsilon with 9J10 peptide | | Descriptor: | 14-3-3 protein epsilon, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9J10 | | Authors: | Mathivanan, S, Sudhakar, S, Bairy, S, Kamariah, N, Venkitaraman, A. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Target identification for small-molecule discovery in the FOXO3a tumor-suppressor pathway using a biodiverse peptide library.
Cell Chem Biol, 28, 2021
|
|
7DDC
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with Tafenoquine | | Descriptor: | 3C-like proteinase, Tafenoquine | | Authors: | Chen, Y, Wang, Y.C, Yang, C.S, Hung, M.C. | | Deposit date: | 2020-10-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Tafenoquine and its derivatives as inhibitors for the severe acute respiratory syndrome coronavirus 2.
J.Biol.Chem., 298, 2022
|
|
6SWO
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH iBET-BD1 (GSK778) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(methoxymethyl)-1-[(1~{R})-1-phenylethyl]-8-[[(3~{S})-pyrrolidin-3-yl]methoxy]imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 2 | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
6SWN
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH iBET-BD1 (GSK778) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(methoxymethyl)-1-[(1~{R})-1-phenylethyl]-8-[[(3~{S})-pyrrolidin-3-yl]methoxy]imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4, ... | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
6SWQ
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH iBET-BD2 (GSK046) | | Descriptor: | 1,2-ETHANEDIOL, 4-acetamido-3-fluoranyl-~{N}-(4-oxidanylcyclohexyl)-5-[(1~{S})-1-phenylethoxy]benzamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
6SWP
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH iBET-BD2 (GSK046) | | Descriptor: | 1,2-ETHANEDIOL, 4-acetamido-3-fluoranyl-~{N}-(4-oxidanylcyclohexyl)-5-[(1~{S})-1-phenylethoxy]benzamide, Bromodomain-containing protein 2 | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
7USI
 
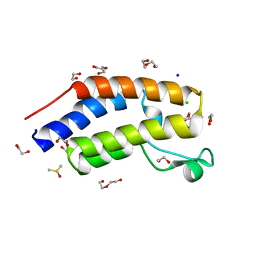 | | BRD2-BD1 in complex with MDP5 | | Descriptor: | (8M)-8-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-(morpholin-4-yl)-4H-1-benzopyran-4-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ... | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7USG
 
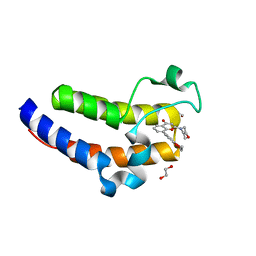 | | BRD2-BD2 in complex with MDP5 | | Descriptor: | (8M)-8-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-(morpholin-4-yl)-4H-1-benzopyran-4-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ... | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7USH
 
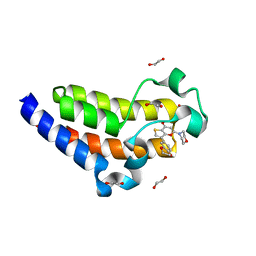 | | BRD2-BD2 in complex with SF2523 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 2 | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7USJ
 
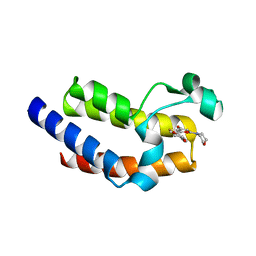 | | BRD4-BD2 in complex with SF2523 | | Descriptor: | 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 4 | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
7USK
 
 | | BRD4-BD2 Ligand free | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
6VOT
 
 | | Crystal structure of Pseudomonas aerugonisa PBP3 complexed to gamma-lactam YU253434 | | Descriptor: | 1-[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-3-oxopropyl]-4-{[2-(5,6 -dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl]carbamoyl}-2,5-dihydro-1H-pyrazole-3-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F. | | Deposit date: | 2020-01-31 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A gamma-Lactam Siderophore Antibiotic Effective against Multidrug-Resistant Gram-Negative Bacilli.
J.Med.Chem., 63, 2020
|
|
1JUG
 
 | | LYSOZYME FROM ECHIDNA MILK (TACHYGLOSSUS ACULEATUS) | | Descriptor: | CALCIUM ION, LYSOZYME | | Authors: | Guss, J.M. | | Deposit date: | 1996-10-13 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the calcium-binding echidna milk lysozyme at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
7LNU
 
 | | Ternary complex of the Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus bound to isopentenyl monophosphate and ATP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7LNW
 
 | |
7LNX
 
 | | I146A mutant of the isopentenyl phosphate kinase from Candidatus methanomethylophilus alvus | | Descriptor: | (2E)-3-methylhept-2-en-1-yl dihydrogen phosphate, (2Z)-3-methylhept-2-en-1-yl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thomas, L.M, Singh, S, Johnson, B.P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7LNT
 
 | | Ternary complex of the Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus bound to benzyl monophosphate and ATP | | Descriptor: | (phenylmethyl) dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
7LNV
 
 | | Apo Structure of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus | | Descriptor: | GLYCEROL, Isopentenyl phosphate kinase, MALONATE ION | | Authors: | Thomas, L.M, Singh, S, Scull, E.M, Bourne, C.R. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for the Substrate Promiscuity of Isopentenyl Phosphate Kinase from Candidatus methanomethylophilus alvus .
Acs Chem.Biol., 17, 2022
|
|
