2P4L
 
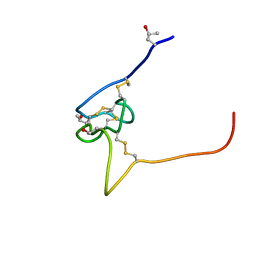 | | Structure and sodium channel activity of an excitatory I1-superfamily conotoxin | | Descriptor: | I-superfamily conotoxin r11a | | Authors: | Buczek, O, Wei, D.X, Babon, J.J, Yang, X.D, Fiedler, B, Yoshikami, D, Olivera, B.M, Bulaj, G, Norton, R.S. | | Deposit date: | 2007-03-12 | | Release date: | 2007-09-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and Sodium Channel Activity of an Excitatory I(1)-Superfamily Conotoxin
Biochemistry, 46, 2007
|
|
6DHL
 
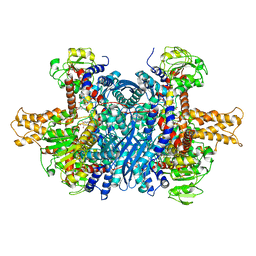 | | Bovine glutamate dehydrogenase complexed with epicatechin-3-gallate (ECG) | | Descriptor: | (2R,3S)-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | Green tea polyphenols control dysregulated glutamate dehydrogenase in transgenic mice by hijacking the ADP activation site.
J. Biol. Chem., 286, 2011
|
|
7D9V
 
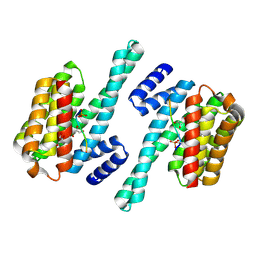 | |
7D8P
 
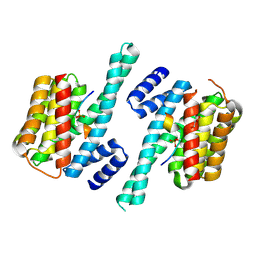 | |
7D8H
 
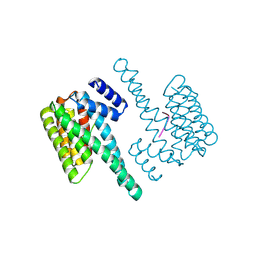 | |
5YHY
 
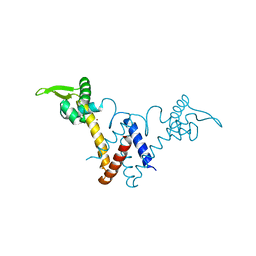 | | Structure of Lactococcus lactis ZitR, C30S mutant | | Descriptor: | ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
5YI3
 
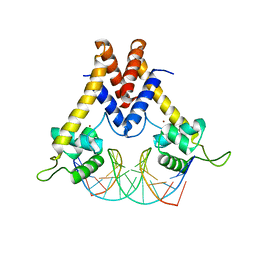 | | Structure of Lactococcus lactis ZitR, C30S mutant in complex with DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*A)-3'), ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
5YHZ
 
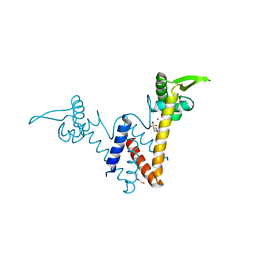 | | Structure of Lactococcus lactis ZitR, E41A mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ZINC ION, ... | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
5YHX
 
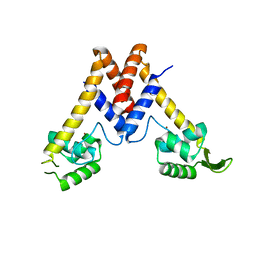 | | Structure of Lactococcus lactis ZitR, wild type | | Descriptor: | ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
5YI0
 
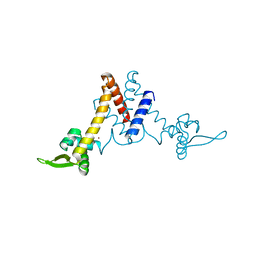 | | Structure of Lactococcus lactis ZitR, C30AH42A mutant | | Descriptor: | ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
5YI1
 
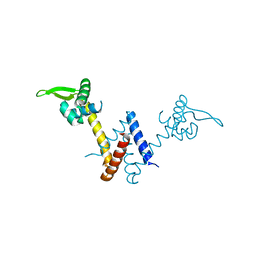 | | Structure of Lactococcus lactis ZitR, C30AH42A mutant in apo form | | Descriptor: | Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
5YI2
 
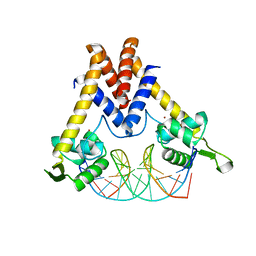 | | Structure of Lactococcus lactis ZitR, wild type in complex with DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*A)-3'), ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
7DMY
 
 | | The crystal structure of Cpd7 in complex with BPTF bromodomain | | Descriptor: | Nucleosome-remodeling factor subunit BPTF, tert-butyl 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-4-oxidanylidene-5,7-dihydropyrrolo[3,4-d]pyrimidine-6-carboxylate | | Authors: | Xiong, L, Guo, Y, Yang, S. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
7DN4
 
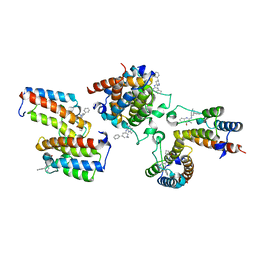 | | The crystal structure of Cpd8 in complex with BPTF bromodomain | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Xiong, L, Guo, Y, Yang, S. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
7Y67
 
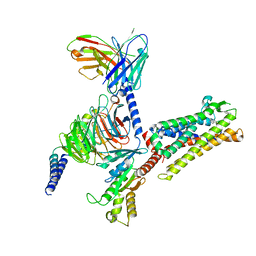 | | Cryo-EM structure of C089-bound C5aR1(I116A) mutant in complex with Gi protein | | Descriptor: | C089 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y65
 
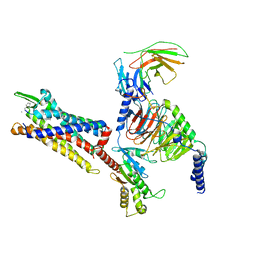 | | Cryo-EM structure of C5a peptide-bound C5aR1 in complex with Gi protein | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, C5apep peptide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y66
 
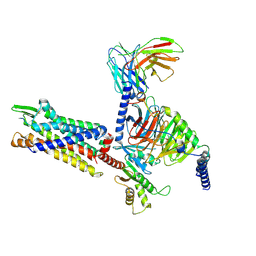 | | Cryo-EM structure of BM213-bound C5aR1 in complex with Gi protein | | Descriptor: | BM213 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y64
 
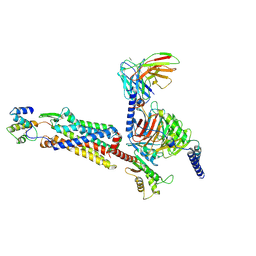 | | Cryo-EM structure of C5a-bound C5aR1 in complex with Gi protein | | Descriptor: | C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
2GAR
 
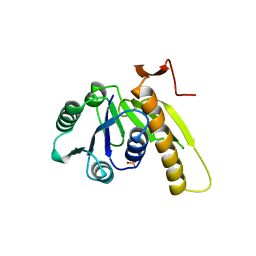 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
7XCD
 
 | |
8HML
 
 | |
7CMD
 
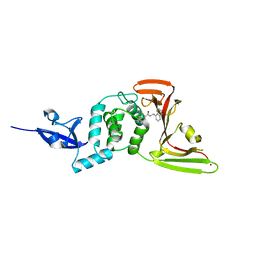 | | Crystal structure of the SARS-CoV-2 PLpro with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease.
Acta Pharm Sin B, 11, 2021
|
|
7CJD
 
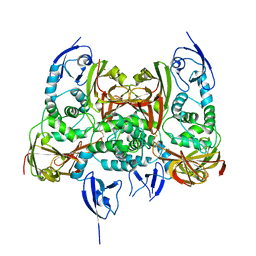 | |
8IFP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 1 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFT
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 10 | | Descriptor: | (8S)-N-[(1S)-1-cyano-2-[(3S)-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2S)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
