6HU2
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 11 | | Descriptor: | 4-methyl-~{N}-oxidanyl-3-[(phenylmethyl)amino]benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HT8
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 3 | | Descriptor: | 3-benzamido-4-methoxy-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
3ESW
 
 | | Complex of yeast PNGase with GlcNAc2-IAc. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase, UV excision repair protein RAD23, ... | | Authors: | Zhao, G, Zhou, X, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and mutational studies on the importance of oligosaccharide binding for the activity of yeast PNGase.
Glycobiology, 19, 2009
|
|
1TLJ
 
 | | Crystal Structure of Conserved Protein of Unknown Function SSO0622 from Sulfolobus solfataricus | | Descriptor: | Hypothetical UPF0130 protein SSO0622, SULFATE ION | | Authors: | Jia, Z, Wong, A.H.Y, Kudrytska, M, Skarina, T, Walker, J, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-09 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional characterization of the TYW3/Taw3 class of SAM-dependent methyltransferases.
Rna, 23, 2017
|
|
6LVK
 
 | | Crystal structure of FGFR2 in complex with 1,3,5-triazine derivative | | Descriptor: | Fibroblast growth factor receptor 2, N-ethyl-2-[[4-[[4-(4-methylpiperazin-1-yl)-3-(2-morpholin-4-ylethoxy)phenyl]amino]-1,3,5-triazin-2-yl]amino]benzenesulfonamide, SULFATE ION | | Authors: | Echizen, Y, Amano, Y, Tateishi, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2.
Bioorg.Med.Chem., 28, 2020
|
|
6N30
 
 | | Bacillus PS3 ATP synthase class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
8JF5
 
 | | Crystal structure of Lysine Specific Demethylase 1 (LSD1) with TAS1440 | | Descriptor: | 4-[5-[(3~{R})-3-azanylpyrrolidin-1-yl]carbonyl-2-[2-fluoranyl-4-(2-methyl-2-oxidanyl-propyl)phenyl]phenyl]-2-fluoranyl-benzenecarbonitrile, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Fukushima, H, Machida, T, Yamashita, S, Suzuki, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | TAS1440, a histone H3 competitive LSD1 inhibitor, activates both TGF-beta and notch signaling pathways via INSM1 dissociation in neuroendocrine small cell lung cancer
To Be Published
|
|
6N2D
 
 | | Bacillus PS3 ATP synthase membrane region | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6N2Z
 
 | | Bacillus PS3 ATP synthase class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6N2Y
 
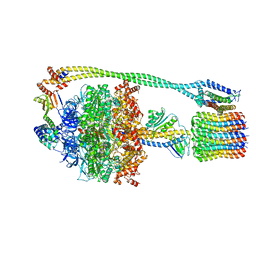 | | Bacillus PS3 ATP synthase class 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6LVM
 
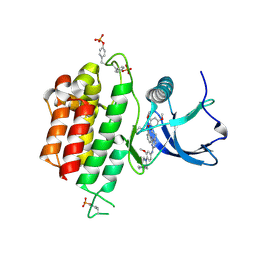 | | Crystal structure of FGFR3 in complex with pyrimidine derivative | | Descriptor: | 2-[[5-[2-(3,5-dimethoxyphenyl)ethyl]-2-[[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]pyrimidin-4-yl]amino]-N-ethyl-benzenesulfonamide, Fibroblast growth factor receptor 3 | | Authors: | Echizen, Y, Tateishi, Y, Amano, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2.
Bioorg.Med.Chem., 28, 2020
|
|
8ZUH
 
 | | Crystal structure of bovine Fbs2/Skp1/Man3GlcNAc2 complex | | Descriptor: | F-box only protein 6, S-phase kinase-associated protein 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Satoh, T, Mizushima, T, Yagi, H, Kato, R, Kato, K. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of sugar recognition by SCF FBS2 ubiquitin ligase involved in NGLY1 deficiency.
Febs Lett., 598, 2024
|
|
6C9N
 
 | |
6C9S
 
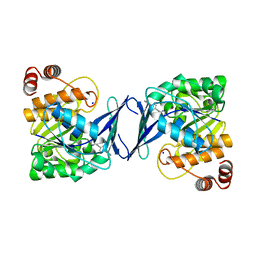 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3R,4S,5R)-2-(6-([1,1'-biphenyl]-4-ylethynyl)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol | | Descriptor: | 6-[([1,1'-biphenyl]-4-yl)ethynyl]-9-beta-D-ribofuranosyl-9H-purine, Adenosine kinase, SODIUM ION, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
6C9R
 
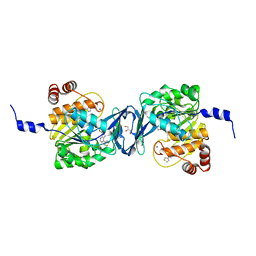 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(thiophen-3-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol | | Descriptor: | 9-beta-D-ribofuranosyl-6-(thiophen-3-yl)-9H-purine, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
6C9Q
 
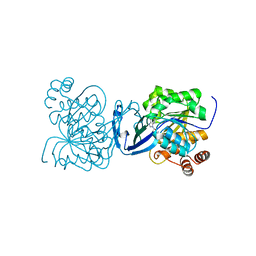 | |
6C9P
 
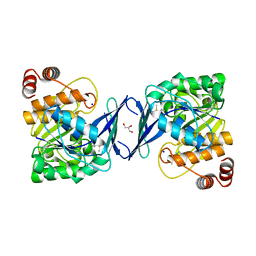 | |
6C67
 
 | | Mycobacterium tuberculosis adenosine kinase bound to iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-17 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
5WXJ
 
 | | Apo EarP | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, GLYCEROL, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXK
 
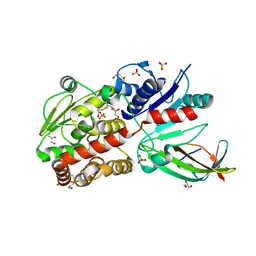 | | EarP bound with domain I of EF-P | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, Elongation factor P, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXI
 
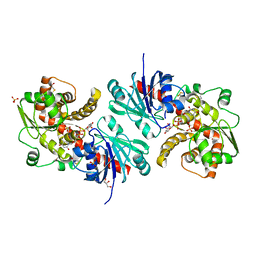 | | EarP bound with dTDP-rhamnose (soaked) | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, BETA-MERCAPTOETHANOL, EarP, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
8JJE
 
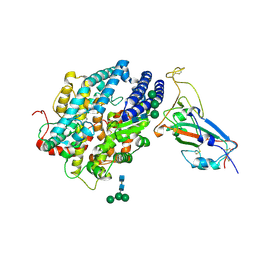 | | RBD of SARS-CoV2 spike protein with ACE2 decoy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kishikawa, J, Hirose, M, Kato, T, Okamoto, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An inhaled ACE2 decoy confers protection against SARS-CoV-2 infection in preclinical models.
Sci Transl Med, 15, 2023
|
|
5XVR
 
 | |
2ELL
 
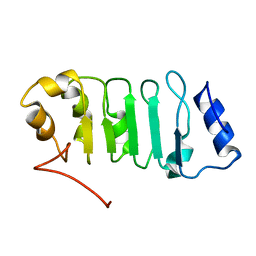 | | Solution structure of the Leucine Rich Repeat of human Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Authors: | Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
5XI8
 
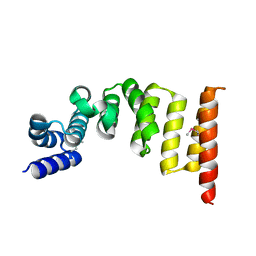 | | Structure and function of the TPR domain | | Descriptor: | Beta-barrel assembly-enhancing protease | | Authors: | Tanaka, Y, Tsukazaki, T. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The TPR domain of BepA is required for productive interaction with substrate proteins and the beta-barrel assembly machinery complex.
Mol. Microbiol., 106, 2017
|
|
