3IML
 
 | |
6CAU
 
 | | UDP-N-acetylmuramate--alanine ligase from Acinetobacter baumannii AB5075-UW with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UDP-N-acetylmuramate--L-alanine ligase | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-01-31 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | UDP-N-acetylmuramate--alanine ligase from Acinetobacter baumannii AB5075-UW with AMPPNP
To be Published
|
|
7LY0
 
 | |
7LXZ
 
 | |
7LY2
 
 | |
7LXX
 
 | |
7LXY
 
 | |
7LY3
 
 | |
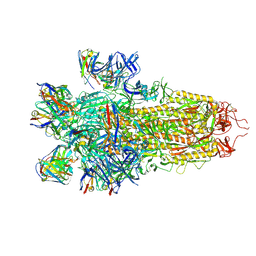
7N8I
 
 | |
7N8H
 
 | |
4PFZ
 
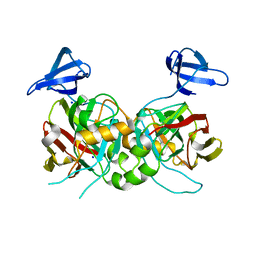 | |
7K4L
 
 | |
7K45
 
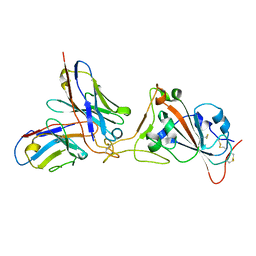 | |
4TMD
 
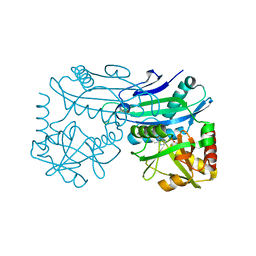
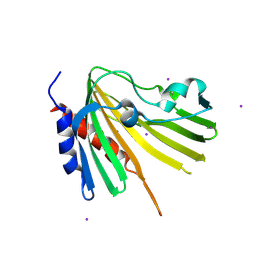 | | X-ray structure of Putative uncharacterized protein (Rv0999 ortholog) from Mycobacterium smegmatis | | Descriptor: | IODIDE ION, Uncharacterized protein | | Authors: | Horanyi, P.S, Dranow, D.M, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-06-01 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Putative uncharacterized protein (Rv0999 ortholog) from Mycobacterium smegmatis
To Be Published
|
|
4TM5
 
 | |
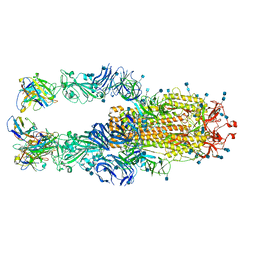
7K4N
 
 | |
7KNA
 
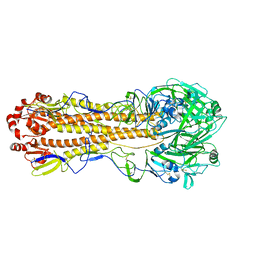 | | Localized reconstruction of the H1 A/Michigan/45/2015 ectodomain displayed at the surface of I53_dn5 nanoparticle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin,I53_dn5 | | Authors: | Acton, O.J, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Quadrivalent influenza nanoparticle vaccines induce broad protection.
Nature, 592, 2021
|
|
4TU1
 
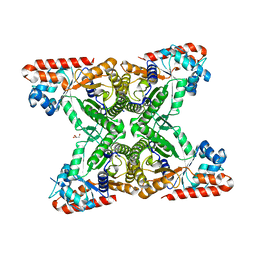 | | Structure of Toxoplasma gondii fructose 1,6 bisphosphate aldolase | | Descriptor: | Fructose-1,6-bisphosphate aldolase, GLYCEROL | | Authors: | Boucher, L.E, Bosch, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-06-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Toxoplasma gondii fructose-1,6-bisphosphate aldolase.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3TL6
 
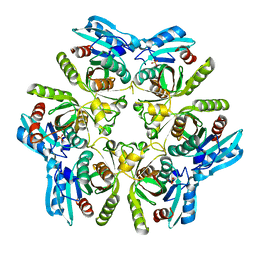 | |
3U40
 
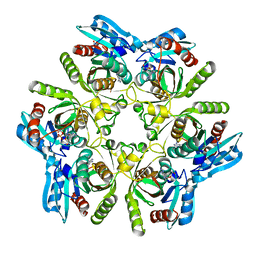 | | Crystal structure of a purine nucleoside phosphorylase from Entamoeba histolytica bound to adenosine | | Descriptor: | ADENOSINE, NITRATE ION, PHOSPHATE ION, ... | | Authors: | Edwards, T.E, Gardberg, A.S, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-10-06 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expression of proteins in Escherichia coli as fusions with maltose-binding protein to rescue non-expressed targets in a high-throughput protein-expression and purification pipeline.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3PME
 
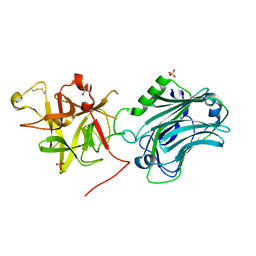 | | Crystal structure of the receptor binding domain of botulinum neurotoxin C/D mosaic serotype | | Descriptor: | GLYCEROL, SULFATE ION, Type C neurotoxin | | Authors: | Zhang, Y, Buchko, G.W, Qin, L, Robinson, H, Varnum, S.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-11-16 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the receptor binding domain of the botulinum C-D mosaic neurotoxin reveals potential roles of lysines 1118 and 1136 in membrane interactions.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
9DF0
 
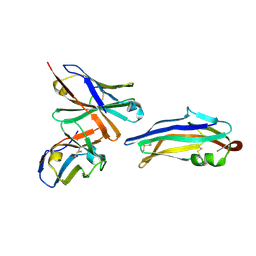 | | PDCoV S RBD bound to PD41 Fab (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PD41 Fab variable heavy-chain, PD41 Fab variable light-chain, ... | | Authors: | Asarnow, D, Rexhepaj, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Isolation and escape mapping of broadly neutralizing antibodies against emerging delta-coronaviruses.
Immunity, 57, 2024
|
|
4PN3
 
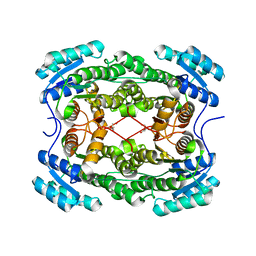 | | Crystal structure of 3-hydroxyacyl-CoA-dehydrogenase from Brucella melitensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyacyl-CoA dehydrogenase | | Authors: | Lukacs, C.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of 3-hydroxyacyl-CoA-dehydrogenase from Brucella melitensis
To Be Published
|
|
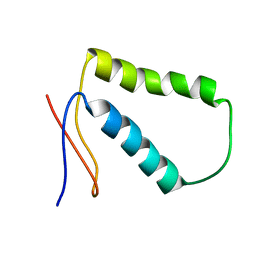
2MRL
 
 | |
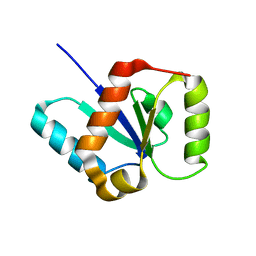
2L2Q
 
 | |
