7UKC
 
 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 1, transmembrane region | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKH
 
 | | Human Kv4.2-KChIP2-DPP6 channel complex in an open state, intracellular region | | Descriptor: | CALCIUM ION, Isoform 2 of Kv channel-interacting protein 2, Potassium voltage-gated channel subfamily D member 2, ... | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKE
 
 | |
7UKG
 
 | | Human Kv4.2-KChIP2-DPP6 channel complex in an open state, transmembrane region | | Descriptor: | Dipeptidyl-peptidase 6, POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UK5
 
 | |
7UKD
 
 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 2, transmembrane region | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKF
 
 | | Human Kv4.2-KChIP2 channel complex in a putative resting state, transmembrane region | | Descriptor: | MERCURY (II) ION, POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
5AYW
 
 | | Structure of a membrane complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Huang, Y, Han, L, Zheng, J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.555 Å) | | Cite: | Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins
Nat.Struct.Mol.Biol., 23, 2016
|
|
5GJA
 
 | | Crystal structure of Arabidopsis thaliana ACO2 in complex with 2-PA | | Descriptor: | 1-aminocyclopropane-1-carboxylate oxidase 2, PYRIDINE-2-CARBOXYLIC ACID, ZINC ION | | Authors: | Sun, X.Z, Li, Y.X, He, W.R, Ji, C.G, Xia, P.X, Wang, Y.C, Du, S, Li, H.J, Raikhel, N, Xiao, J.Y, Guo, H.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase.
Nat Commun, 8, 2017
|
|
6KJY
 
 | | Galectin-13 variant C136S/C138S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
6KJW
 
 | | Galectin-13 variant C136S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
4JH0
 
 | | Crystal structure of dipeptidyl-peptidase 4 (CD26, adenosine deaminase complexing protein 2) (DPP-IV-WT) complex with bms-767778 AKA 2-(3-(aminomethyl)-4-(2,4- dichlorophenyl)-2-methyl-5-oxo-5,7-dihydro-6h-pyrrolo[3,4- b]pyridin-6-yl)-n,n-dimethylacetamide | | Descriptor: | 2-[3-(aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5,7-dihydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-N,N-dimethylacetamide, Dipeptidyl peptidase 4 | | Authors: | Klei, H.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-09-04 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of Activity, Selectivity, and Liability Profiles in 5-Oxopyrrolopyridine DPP4 Inhibitors Leading to Clinical Candidate (Sa)-2-(3-(Aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5H-pyrrolo[3,4-b]pyridin-6(7H)-yl)-N,N-dimethylacetamide (BMS-767778).
J.Med.Chem., 56, 2013
|
|
5HN7
 
 | |
5HNA
 
 | |
6KJX
 
 | | Galectin-13 variant C138S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
5HN8
 
 | | Crystal structure of Plasmodium vivax geranylgeranylpyrophosphate synthase complexed with BPH-1182 | | Descriptor: | 2-{[3-hydroxy-5-(octyloxy)benzyl]sulfanyl}benzoic acid, Farnesyl pyrophosphate synthase, putative, ... | | Authors: | Liu, Y.-L, Zhang, Y, Oldfield, E. | | Deposit date: | 2016-01-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dynamic Structure and Inhibition of a Malaria Drug Target: Geranylgeranyl Diphosphate Synthase.
Biochemistry, 55, 2016
|
|
5HN9
 
 | |
6KXS
 
 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the ectodomain of pIgR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Li, Y, Wang, G, Xiao, J. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-05 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into immunoglobulin M.
Science, 367, 2020
|
|
6KPP
 
 | | BNC105 in complex with tubulin | | Descriptor: | (6-methoxy-2-methyl-7-oxidanyl-1-benzofuran-3-yl)-(3,4,5-trimethoxyphenyl)methanone, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, T, Wu, C, Pu, D. | | Deposit date: | 2019-08-15 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74524117 Å) | | Cite: | Unraveling the molecular mechanism of BNC105, a phase II clinical trial vascular disrupting agent, provides insights into drug design.
Biochem.Biophys.Res.Commun., 2020
|
|
5XLZ
 
 | |
2DHO
 
 | | Crystal structure of human IPP isomerase I in space group P212121 | | Descriptor: | CHLORIDE ION, Isopentenyl-diphosphate delta-isomerase 1, MANGANESE (II) ION, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-03-24 | | Release date: | 2007-06-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
7CJT
 
 | | Crystal Structure of SETDB1 Tudor domain in complexed with (R,R)-59 | | Descriptor: | 2-[[(3~{R},5~{R})-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-3-prop-2-enyl-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y.P, Liang, X, Mao, X, Wu, C, Luyi, H, Yang, S. | | Deposit date: | 2020-07-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.474 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7CAJ
 
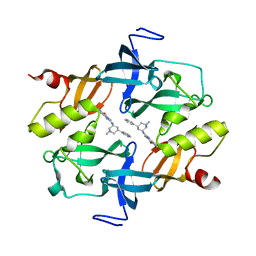 | | Crystal structure of SETDB1 Tudor domain in complexed with Compound 2. | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y.P, Liang, X, Xin, M, Luyi, H, Chengyong, W, Yang, S.Y. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7S0J
 
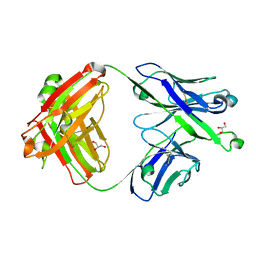 | | Crystal structure of Epstein-Barr virus gH/gL targeting antibody 769B10 | | Descriptor: | 769B10 Fab Heavy chain, 769B10 Fab Light chain, GLYCEROL | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S08
 
 | |
