2DCZ
 
 | | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase By Directed Evolution | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Endo-1,4-beta-xylanase A, SULFATE ION | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2DCY
 
 | | Crystal structure of Bacillus subtilis family-11 xylanase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D(-)-TARTARIC ACID, Endo-1,4-beta-xylanase A, ... | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2E1C
 
 | | Structure of Putative HTH-type transcriptional regulator PH1519/DNA Complex | | Descriptor: | DNA (5'-D(*DAP*DGP*DTP*DGP*DAP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DTP*DCP*DAP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DTP*DGP*DAP*DAP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DCP*DAP*DCP*DT)-3'), Putative HTH-type transcriptional regulator PH1519 | | Authors: | Koike, H, Suzuki, M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-12-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Feast/Famine Regulation by Transcription Factor FL11 for the Survival of the Hyperthermophilic Archaeon Pyrococcus OT3.
Structure, 15, 2007
|
|
2E5A
 
 | | Crystal Structure of Bovine Lipoyltransferase in Complex with Lipoyl-AMP | | Descriptor: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, ACETIC ACID, Lipoyltransferase 1, ... | | Authors: | Fujiwara, K, Hosaka, H, Matsuda, M, Suzuki, M, Nakagawa, A. | | Deposit date: | 2006-12-19 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bovine Lipoyltransferase in complex with lipoyl-AMP
J.Mol.Biol., 371, 2007
|
|
5AZ0
 
 | | Crystal structure of aldo-keto reductase (AKR2E5) of the silkworm, Bombyx mori | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5AZ1
 
 | | Crystal structure of aldo-keto reductase (AKR2E5) complexed with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5B22
 
 | | Dimer structure of murine Nectin-3 D1D2 | | Descriptor: | Nectin-3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Takebe, K, Sangawa, T, Katsutani, T, Narita, H, Suzuki, M. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Dimer structure of murine Nectin-3 D1D2
To Be Published
|
|
5B21
 
 | |
2E0Z
 
 | | Crystal structure of virus-like particle from Pyrococcus furiosus | | Descriptor: | Virus-like particle | | Authors: | Akita, F, Chong, K.T, Tanaka, H, Yamashita, E, Miyazaki, N, Nakaishi, Y, Namba, K, Ono, Y, Suzuki, M, Tsukihara, T, Nakagawa, A. | | Deposit date: | 2006-10-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Crystal Structure of a Virus-like Particle from the Hyperthermophilic Archaeon Pyrococcus furiosus Provides Insight into the Evolution of Viruses
J.Mol.Biol., 368, 2007
|
|
2HI7
 
 | | Crystal structure of DsbA-DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Murakami, S, Suzuki, M, Nakagawa, A, Yamashita, E, Okada, K, Ito, K. | | Deposit date: | 2006-06-29 | | Release date: | 2006-12-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation
Cell(Cambridge,Mass.), 127, 2006
|
|
8K6T
 
 | | The minor pilin structure of FctB3 in Streptococcus | | Descriptor: | FctB3, GLYCEROL | | Authors: | Takebe, K, Sangawa, T, Suzuki, M, Nakata, M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of FctB3 crystal structure and insight into its structural stabilization and pilin linkage mechanisms.
Arch.Microbiol., 206, 2023
|
|
8JZ8
 
 | | Subatomic structure of orthorhombic thaumatin at 0.89 Angstroms | | Descriptor: | DI(HYDROXYETHYL)ETHER, Thaumatin I | | Authors: | Masuda, T, Suzuki, M, Yamasaki, M, Mikami, B. | | Deposit date: | 2023-07-04 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Subatomic structure of orthorhombic thaumatin at 0.89 angstrom reveals that highly flexible conformations are crucial for thaumatin sweetness.
Biochem.Biophys.Res.Commun., 703, 2024
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7D5M
 
 | | Crystal structure of inositol dehydrogenase homolog complexed with NAD+ from Azotobacter vinelandii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase | | Authors: | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of inositol dehydrogenase complexed with NAD+ from Azotobacter vinelandii
To Be Published
|
|
7D5N
 
 | | Crystal structure of inositol dehydrogenase homolog complexed with NADH and myo-inositol from Azotobacter vinelandii | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of inositol dehydrogenase complexed with NADH and myo-inositol from Azotobacter vinelandii
To Be Published
|
|
7CMM
 
 | | Crystal structure of TEAD1-YBD in complex with K-975 | | Descriptor: | N-[3-(4-chloranylphenoxy)-4-methyl-phenyl]propanamide, Transcriptional enhancer factor TEF-1 | | Authors: | Tsuji, Y, Suzuki, M, Yasunaga, M, Hamguchi, K, Saito, J. | | Deposit date: | 2020-07-28 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The novel potent TEAD inhibitor, K-975, inhibits YAP1/TAZ-TEAD protein-protein interactions and exerts an anti-tumor effect on malignant pleural mesothelioma.
Am J Cancer Res, 10, 2020
|
|
2Z5P
 
 | | Apo-Fr with low content of Pd ions | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Ueno, T, Hirata, K, Abe, M, Suzuki, M, Abe, S, Shimizu, N, Yamamoto, M, Takata, M, Watanabe, Y. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Process of accumulation of metal ions on the interior surface of apo-ferritin: crystal structures of a series of apo-ferritins containing variable quantities of Pd(II) ions.
J.Am.Chem.Soc., 131, 2009
|
|
2Z5R
 
 | | Apo-Fr with high content of Pd ions | | Descriptor: | CADMIUM ION, Ferritin light chain, PALLADIUM ION | | Authors: | Ueno, T, Hirata, K, Abe, M, Suzuki, M, Abe, S, Shimizu, N, Yamamoto, M, Takata, M, Watanabe, Y. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Process of accumulation of metal ions on the interior surface of apo-ferritin: crystal structures of a series of apo-ferritins containing variable quantities of Pd(II) ions.
J.Am.Chem.Soc., 131, 2009
|
|
2Z5Q
 
 | | Apo-Fr with intermediate content of Pd ion | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Ueno, T, Hirata, K, Abe, M, Suzuki, M, Abe, S, Shimizu, N, Yamamoto, M, Takata, M, Watanabe, Y. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Process of accumulation of metal ions on the interior surface of apo-ferritin: crystal structures of a series of apo-ferritins containing variable quantities of Pd(II) ions.
J.Am.Chem.Soc., 131, 2009
|
|
3ASX
 
 | | Human Squalene synthase in complex with 1-{4-[{4-chloro-2-[(2-chlorophenyl)(hydroxy)methyl]phenyl}(2,2-dimethylpropyl)amino]-4-oxobutanoyl}piperidine-3-carboxylic acid | | Descriptor: | (3R)-1-{4-[{4-chloro-2-[(S)-(2-chlorophenyl)(hydroxy)methyl]phenyl}(2,2-dimethylpropyl)amino]-4-oxobutanoyl}piperidine-3-carboxylic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Shimizu, H, Suzuki, M, Katakura, S, Yamazaki, K, Higashihashi, N, Ichikawa, M, Yokomizo, A, Itoh, M, Sugita, K, Usui, H. | | Deposit date: | 2010-12-22 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a new 2-aminobenzhydrol template for highly potent squalene synthase inhibitors
Bioorg.Med.Chem., 19, 2011
|
|
2Z4P
 
 | | Crystal structure of FFRP-DM1 | | Descriptor: | 75aa long hypothetical regulatory protein AsnC, ISOLEUCINE | | Authors: | Yamada, M, Koike, H, Kudo, N, Suzuki, M. | | Deposit date: | 2007-06-21 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Structural Code for Discriminating between Transcription Signals Revealed by the Feast/Famine Regulatory Protein DM1 in Complex with Ligands
Structure, 15, 2007
|
|
7DDA
 
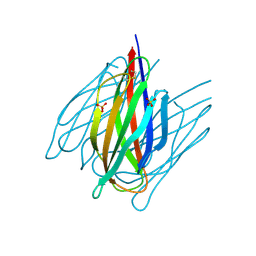 | | Envelope protein VP37 a crystal structure from White Spot Syndrome Virus | | Descriptor: | Envelope protein, SULFATE ION | | Authors: | Somsoros, W, Sangawa, T, Takebe, K, Attarataya, J, Suzuki, M, Khunrae, P. | | Deposit date: | 2020-10-28 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of the C-terminal domain of envelope protein VP37 from white spot syndrome virus reveals sulphate binding sites responsible for heparin binding.
J.Gen.Virol., 102, 2021
|
|
3VWU
 
 | |
3WUM
 
 | |
3VWV
 
 | |
