7AIX
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide | | Descriptor: | 2-{1-[4-(12-Amino-3-chloro-6,7,10,11-tetrahydro-7,11-methanocycloocta[b]quinolin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}-N-[4-hydroxy-3-methoxybenzyl]acetamide, Acetylcholinesterase, CHLORIDE ION, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
7AIW
 
 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with (E)-10-[(3-Chloro-6,7,10,11-tetrahydro-9-methyl-7,11-methanocycloocta[b]quinolin-12-yl)amino]-N-(4-hydroxy-3-methoxybenzyl)-6-decenamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Potent Dual Inhibitor of Acetylcholinesterase and Butyrylcholinesterase with Antioxidant Activity that Alleviates Alzheimer-like Pathology in Old APP/PS1 Mice.
J.Med.Chem., 64, 2021
|
|
4FGW
 
 | | Structure of Glycerol-3-Phosphate Dehydrogenase, GPD1, from Sacharomyces Cerevisiae | | Descriptor: | Glycerol-3-phosphate dehydrogenase [NAD(+)] 1 | | Authors: | Aparicio, D, Munmun, N, Carpena, X, Fita, I, Loewen, P. | | Deposit date: | 2012-06-05 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase (GPD1) from Saccharomyces cerevisiae at 2.45A resolution
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4BB4
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N-(2-methoxyethyl)-4-[(6-pyridin-4-ylquinazolin-2-yl)amino]benzamide | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Optimization of a Novel Series of Potent Mutant B-Raf V600E Selective Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4AYB
 
 | | RNAP at 3.2Ang | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Wojtas, M.N, Mogni, M, Millet, O, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural and Functional Analyses of the Interaction of Archaeal RNA Polymerase with DNA.
Nucleic Acids Res., 40, 2012
|
|
2QE2
 
 | | Structure of HCV NS5B Bound to an Anthranilic Acid Inhibitor | | Descriptor: | 2-{[N-(2-ACETYL-5-CHLORO-4-FLUOROPHENYL)GLYCYL]AMINO}BENZOIC ACID, RNA-directed RNA polymerase | | Authors: | Chopra, R, Svenson, K, Bard, J. | | Deposit date: | 2007-06-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of Anthranilic Acid Derivatives as a Novel Class of Allosteric Inhibitors of Hepatitis C NS5B Polymerase
J.Med.Chem., 50, 2007
|
|
2QE5
 
 | |
2MCR
 
 | | Solution structure of ShK-like immunomodulatory peptide from Brugia malayi (filarial worm) | | Descriptor: | Probable zinc metalloproteinase, putative | | Authors: | Chhabra, S, Swarbrick, J.D, Pennington, M.W, Chang, S.C, Norton, R.S. | | Deposit date: | 2013-08-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Kv1.3 channel-blocking immunomodulatory peptides from parasitic worms: implications for autoimmune diseases.
Faseb J., 28, 2014
|
|
2MD0
 
 | | Solution structure of ShK-like immunomodulatory peptide from Ancylostoma caninum (hookworm) | | Descriptor: | AcK1 | | Authors: | Chhabra, S, Swarbrick, J.D, Mohanty, B, Chang, S.C, Chandy, G.K, Pennington, M.W, Norton, R.S. | | Deposit date: | 2013-08-28 | | Release date: | 2014-07-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Kv1.3 channel-blocking immunomodulatory peptides from parasitic worms: implications for autoimmune diseases.
Faseb J., 28, 2014
|
|
3PIK
 
 | | Outer membrane protein CusC | | Descriptor: | Cation efflux system protein cusC, SULFATE ION, UNKNOWN LIGAND | | Authors: | van den Berg, B. | | Deposit date: | 2010-11-06 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli CusC, the Outer Membrane Component of a Heavy Metal Efflux Pump.
Plos One, 6, 2011
|
|
3PGS
 
 | | Phe3Gly mutant of EcFadL | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ACETATE ION, CALCIUM ION, ... | | Authors: | van den Berg, B, Lepore, B.W. | | Deposit date: | 2010-11-02 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Cover: Ligand-gated diffusion across the bacterial outer membrane.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2JAI
 
 | | DDAH1 complexed with citrulline | | Descriptor: | CITRULLINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Murray-Rust, J, O'Hara, B.P, Rossiter, S, Leiper, J.M, Vallance, P, McDonald, N.Q. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of methylarginine metabolism impairs vascular homeostasis.
Nat. Med., 13, 2007
|
|
2JAJ
 
 | | DDAH1 complexed with L-257 | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1, N~5~-{IMINO[(2-METHOXYETHYL)AMINO]METHYL}-L-ORNITHINE | | Authors: | Murray-Rust, J, O'Hara, B.P, Rossiter, S, Leiper, J.M, Vallance, P, McDonald, N.Q. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disruption of methylarginine metabolism impairs vascular homeostasis.
Nat. Med., 13, 2007
|
|
2YDK
 
 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 2-(CARBAMOYLAMINO)-5-PHENYL-N-[(3S)-PIPERIDIN-3-YL]THIOPHENE-3-CARBOXAMIDE, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
1RFK
 
 | |
2YEX
 
 | | Synthesis and evaluation of triazolones as checkpoint kinase 1 inhibitors | | Descriptor: | 5-METHYL-8-(1H-PYRROL-2-YL)[1,2,4]TRIAZOLO[4,3-A]QUINOLIN-1(2H)-ONE, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Vallentine, A, White, A. | | Deposit date: | 2011-03-31 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis and Evaluation of Triazolones as Checkpoint Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2YER
 
 | | Synthesis and evaluation of triazolones as checkpoint kinase 1 inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 5-(HYDROXYMETHYL)-8-(1H-PYRROL-2-YL)-2H-[1,2,4]TRIAZOLO[4,3-A]QUINOLIN-1-ONE, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Vallentine, A, White, A, Otterbein, L. | | Deposit date: | 2011-03-30 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Synthesis and Evaluation of Triazolones as Checkpoint Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2YDJ
 
 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 5-(3-fluorophenyl)-N-[(3S)-3-piperidyl]-3-ureido-thiophene-2-carboxamide, PHOSPHATE ION, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-22 | | Release date: | 2012-01-25 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
2YDI
 
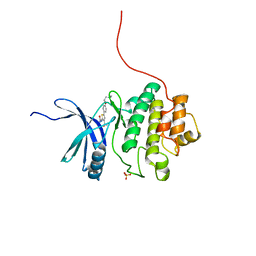 | | Discovery of Checkpoint Kinase Inhibitor AZD7762 by Structure Based Design and Optimization of Thiophene Carboxamide Ureas | | Descriptor: | 5-[4-(2-DIMETHYLAMINOETHYLOXY)PHENYL]-2-UREIDO-THIOPHENE-3-CARBOXAMIDE, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Rowsell, S, Vallentine, A, White, A. | | Deposit date: | 2011-03-21 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(Piperidin-3-Yl)-3-Ureidothiophene-2-Carboxamide (Azd7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
2GNA
 
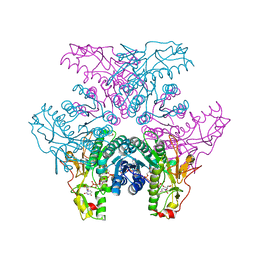 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP-Gal | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GALACTOSE-URIDINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
2FST
 
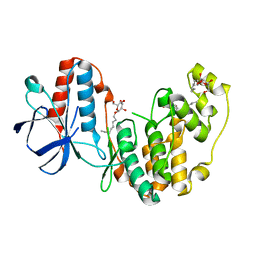 | |
2FSM
 
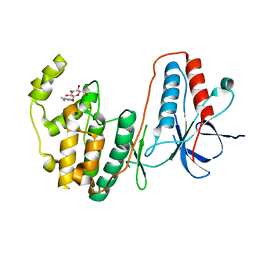 | |
2FSO
 
 | |
2FSL
 
 | |
2GN8
 
 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
