5CTV
 
 | | Catalytic domain of LytA, the major autolysin of Streptococcus pneumoniae, (C60A, H133A, C136A mutant) complexed with peptidoglycan fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, Autolysin, fragment of peptidoglycan | | Authors: | Achour, A, Sandalova, T, Mellroth, P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of the major pneumococcal autolysin LytA in complex with a large peptidoglycan fragment reveals the pivotal role of glycans for lytic activity.
Mol.Microbiol., 101, 2016
|
|
6JN8
 
 | | Structure of H216A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, SULFATE ION, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMZ
 
 | | Structure of H247A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMX
 
 | | Structure of open form of peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | Descriptor: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
5OIZ
 
 | | Penicillin-Binding Protein 2X (PBP2X) from Streptococcus pneumoniae in complex with oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 2X | | Authors: | Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allostery, Recognition of Nascent Peptidoglycan, and Cross-linking of the Cell Wall by the Essential Penicillin-Binding Protein 2x of Streptococcus pneumoniae.
ACS Chem. Biol., 13, 2018
|
|
6JN0
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | Descriptor: | C0O-DAL-API, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
 | | Structure of H216A mutant closed form peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMY
 
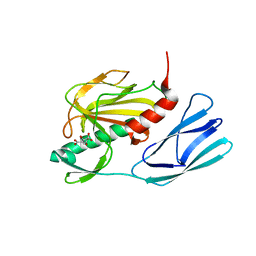 | | Structure of wild type closed form of peptidoglycan peptidase | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6I0N
 
 | |
5OJ1
 
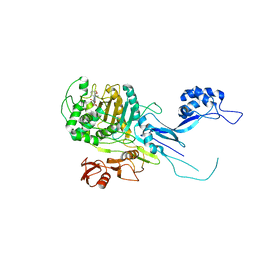 | | Penicillin Binding Protein 2x (PBP2x) from S.pneumoniae in complex with Oxacillin and a tetrasaccharide | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 2X, SODIUM ION | | Authors: | Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allostery, Recognition of Nascent Peptidoglycan, and Cross-linking of the Cell Wall by the Essential Penicillin-Binding Protein 2x of Streptococcus pneumoniae.
ACS Chem. Biol., 13, 2018
|
|
5O8X
 
 | |
6I09
 
 | |
6I0A
 
 | |
5OJ0
 
 | |
6KV1
 
 | | Structure of wild type closed form of peptidoglycan peptidase ZN SAD | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
5OM9
 
 | | Crystal structure of the human CARBOXYPEPTIDASE A1 in complex with a thiirane mechanism-based inhibitor | | Descriptor: | (2~{R})-4-methyl-2-[(1~{S})-1-sulfanylethyl]pentanoic acid, Carboxypeptidase A1, ZINC ION | | Authors: | Gallego, P, Granados, C, Fernandez, D, Pallares, I, Covaleda, G, Aviles, F.X, Vendrell, J, Reverter, D. | | Deposit date: | 2017-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Mechanism-Based Inactivators for Human Pancreatic Carboxypeptidase A from a Focused Synthetic Library.
ACS Med Chem Lett, 8, 2017
|
|
6I05
 
 | |
4CPK
 
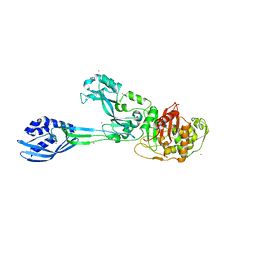 | | Crystal structure of PBP2a double clinical mutant N146K-E150K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, Penicillin binding protein 2 prime, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
4CHX
 
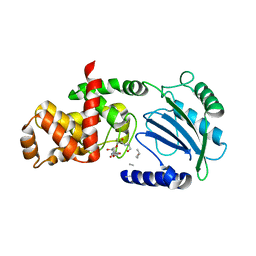 | | Crystal structure of MltC in complex with disaccharide pentapeptide DHl89 | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-bound lytic murein transglycosylase C, ... | | Authors: | Artola-Recolons, C, Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2013-12-04 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase Mltc of Escherichia Coli.
Acs Chem.Biol., 9, 2014
|
|
4JF4
 
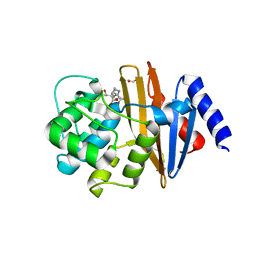 | | OXA-23 meropenem complex | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B, Toth, M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-25 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Basis for Carbapenemase Activity of the OXA-23 beta-Lactamase from Acinetobacter baumannii.
Chem.Biol., 20, 2013
|
|
4JF6
 
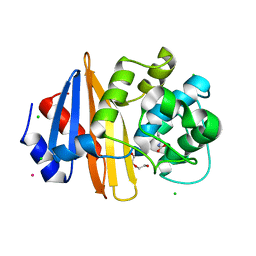 | | Structure of OXA-23 at pH 7.0 | | Descriptor: | Beta-lactamase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-25 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Carbapenemase Activity of the OXA-23 beta-Lactamase from Acinetobacter baumannii.
Chem.Biol., 20, 2013
|
|
4JF5
 
 | | Structure of OXA-23 at pH 4.1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Basis for Carbapenemase Activity of the OXA-23 beta-Lactamase from Acinetobacter baumannii.
Chem.Biol., 20, 2013
|
|
4GNU
 
 | | Crystal structure of GES-5 carbapenemase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase GES-5 | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2012-08-17 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structural basis for progression toward the carbapenemase activity in the GES family of beta-lactamases.
J.Am.Chem.Soc., 134, 2012
|
|
4H8R
 
 | | Imipenem complex of GES-5 carbapenemase | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Extended-spectrum beta-lactamase GES-5, IODIDE ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2012-09-23 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for progression toward the carbapenemase activity in the GES family of beta-lactamases.
J.Am.Chem.Soc., 134, 2012
|
|
