1IEU
 
 | |
1ID2
 
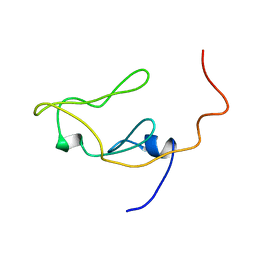
 | | CRYSTAL STRUCTURE OF AMICYANIN FROM PARACOCCUS VERSUTUS (THIOBACILLUS VERSUTUS) | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Romero, A, Nar, H, Messerschmidt, A. | | Deposit date: | 2001-04-03 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure analysis and refinement at 2.15 A resolution of amicyanin, a type I blue copper protein, from Thiobacillus versutus.
J.Mol.Biol., 236, 1994
|
|
3R3G
 
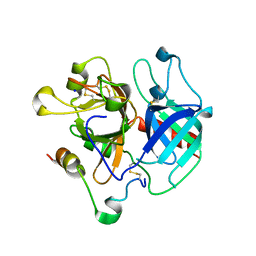 | | Structure of human thrombin with residues 145-150 of murine thrombin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Thrombin Heavy Chain, ... | | Authors: | Pozzi, N, Chen, R, Chen, Z, Bah, A, Di Cera, E. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rigidification of the autolysis loop enhances Na(+) binding to thrombin.
Biophys.Chem., 159, 2011
|
|
3S7H
 
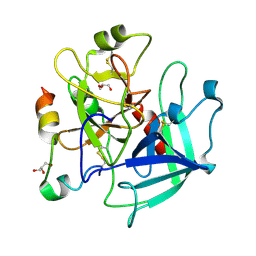 | | Structure of thrombin mutant Y225P in the E* form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Prothrombin | | Authors: | Niu, W, Chen, Z, Gandhi, P, Vogt, A, Pozzi, N, Pele, L.A, Zapata, F, Di Cera, E. | | Deposit date: | 2011-05-26 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Kinetic Evidence of Allostery in a Trypsin-like Protease.
Biochemistry, 50, 2011
|
|
3QDZ
 
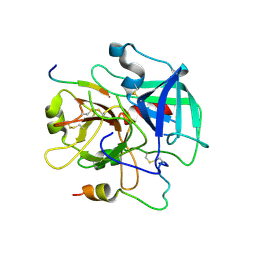 | | Crystal structure of the human thrombin mutant D102N in complex with the extracellular fragment of human PAR4. | | Descriptor: | Proteinase-activated receptor 4, Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P, Chen, Z, Appelbaum, E, Zapata, F, Di Cera, E. | | Deposit date: | 2011-01-19 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of thrombin-protease-receptor interactions
IUBMB LIFE, 63, 2011
|
|
3S7K
 
 | | Structure of thrombin mutant Y225P in the E form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, POTASSIUM ION, Prothrombin | | Authors: | Niu, W, Chen, Z, Gandhi, P, Vogt, A, Pozzi, N, Pele, L.A, Zapata, F, Di Cera, E. | | Deposit date: | 2011-05-26 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Kinetic Evidence of Allostery in a Trypsin-like Protease.
Biochemistry, 50, 2011
|
|
2MAD
 
 | |
1LTD
 
 | |
