8TL2
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-c.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TL3
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-d.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP glycoprotein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TL5
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-c.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TKC
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-b.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TL4
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-e.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
7TBF
 
 | | Locally refined region of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SARS-CoV-2 antibody A19-61.1, Heavy chain of SARS-CoV-2 antibody B1-182.1, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7TB8
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7TCA
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibody A19-46.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody A19-46.1, ... | | Authors: | Zhou, T, Kwong, P.D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7TCC
 
 | |
7TC9
 
 | |
7U0D
 
 | |
8EUV
 
 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
8ELI
 
 | |
8EUU
 
 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
8EUW
 
 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
8WCF
 
 | | Crystal structure of EcThsB | | Descriptor: | Molecular chaperone Tir, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Chen, Q, Yu, Y. | | Deposit date: | 2023-09-12 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Self-association activates ThsB NAD + hydrolase for defense against phage infection.
Biochem.Biophys.Res.Commun., 776, 2025
|
|
8WC0
 
 | | Crystal structure of EcThsA | | Descriptor: | GLYCEROL, NAD(+) hydrolase ThsA, SODIUM ION, ... | | Authors: | Chen, Q, Yu, Y. | | Deposit date: | 2023-09-11 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Self-association activates ThsB NAD + hydrolase for defense against phage infection.
Biochem.Biophys.Res.Commun., 776, 2025
|
|
8FJZ
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(3R,5S)-3-Amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | Descriptor: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP3
 
 | | PKCeta kinase domain in complex with compound 11 | | Descriptor: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Protein kinase C eta type | | Authors: | Johnson, E. | | Deposit date: | 2023-01-03 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP1
 
 | | PKCeta kinase domain in complex with compound 2 | | Descriptor: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Protein kinase C eta type | | Authors: | Johnson, E. | | Deposit date: | 2023-01-03 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FH4
 
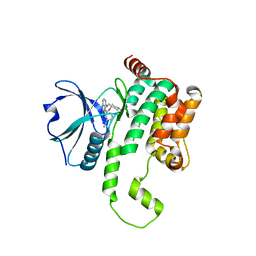 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undec-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile | | Descriptor: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-13 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FKO
 
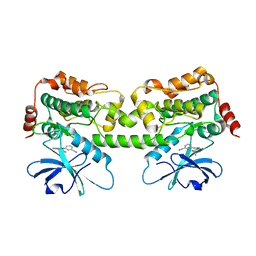 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(2S,5R)-5-Amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | Descriptor: | (3P)-3-{4-[(2S,5R)-5-amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-21 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
3R1B
 
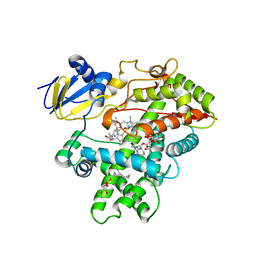 | | Open crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
3UAS
 
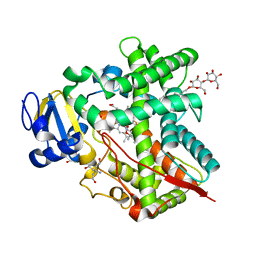 | | Cytochrome P450 2B4 covalently bound to the mechanism-based inactivator 9-ethynylphenanthrene | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gay, S.C, Zhang, H, Shah, M.B, Stout, C.D, Halpert, J.R, Hollenberg, P.F. | | Deposit date: | 2011-10-21 | | Release date: | 2013-01-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | Potent Mechanism-Based Inactivation of Cytochrome P450 2B4 by 9-Ethynylphenanthrene: Implications for Allosteric Modulation of Cytochrome P450 Catalysis.
Biochemistry, 52, 2013
|
|
3R1A
 
 | | Closed crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
