8FJZ
 
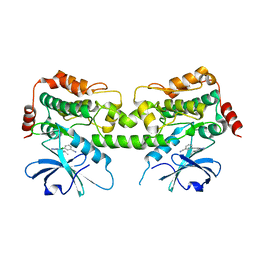 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(3R,5S)-3-Amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | Descriptor: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP3
 
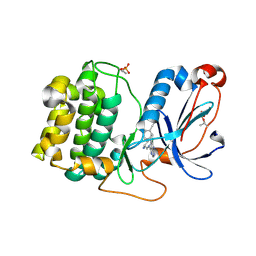 | | PKCeta kinase domain in complex with compound 11 | | Descriptor: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Protein kinase C eta type | | Authors: | Johnson, E. | | Deposit date: | 2023-01-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FH4
 
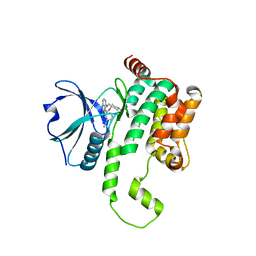 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undec-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile | | Descriptor: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-13 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FKO
 
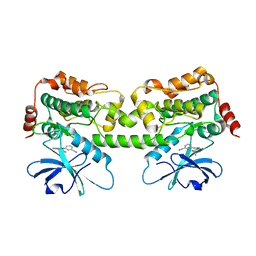 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(2S,5R)-5-Amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | Descriptor: | (3P)-3-{4-[(2S,5R)-5-amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-21 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP1
 
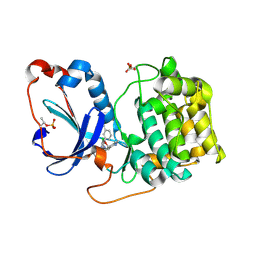 | | PKCeta kinase domain in complex with compound 2 | | Descriptor: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Protein kinase C eta type | | Authors: | Johnson, E. | | Deposit date: | 2023-01-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
5ZG9
 
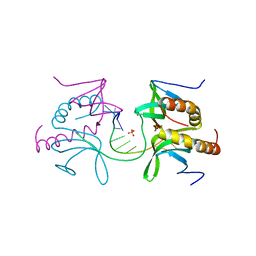 | | Crystal structure of MoSub1-ssDNA complex in phosphate buffer | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*G)-3'), MoSub1, PHOSPHATE ION | | Authors: | Zhao, Y, Huang, J, Liu, H, Yi, L, Wang, S, Zhang, X, Liu, J. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The effect of phosphate ion on the ssDNA binding mode of MoSub1, a Sub1/PC4 homolog from rice blast fungus.
Proteins, 87, 2019
|
|
4FJQ
 
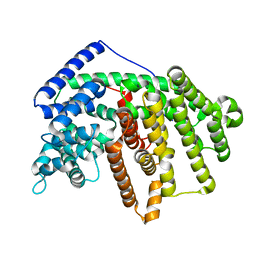 | | Crystal Structure of an alpha-Bisabolol synthase | | Descriptor: | Amorpha-4,11-diene synthase | | Authors: | Jianxu, L, Peng, Z. | | Deposit date: | 2012-06-12 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0001 Å) | | Cite: | Rational engineering of plasticity residues of sesquiterpene synthases from Artemisia annua: product specificity and catalytic efficiency.
Biochem.J., 451, 2013
|
|
3US9
 
 | | Crystal Structure of the NCX1 Intracellular Tandem Calcium Binding Domains(CBD12) | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1 | | Authors: | Giladi, M, Sasson, Y, Hirsch, J.A, Khananshvili, D. | | Deposit date: | 2011-11-23 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A common Ca2+-driven interdomain module governs eukaryotic NCX regulation.
Plos One, 7, 2012
|
|
7EMN
 
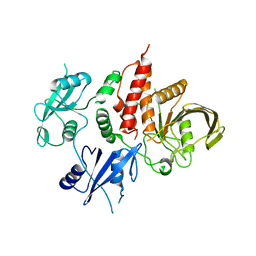 | | The atomic structure of SHP2 E76A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, F, Xie, J.J, Zhu, J.D, Liu, C. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel partially open state of SHP2 points to a "multiple gear" regulation mechanism.
J.Biol.Chem., 296, 2021
|
|
4M1P
 
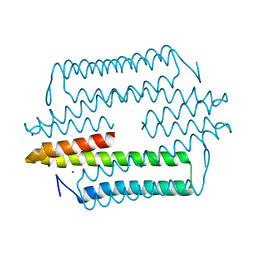 | |
7AVQ
 
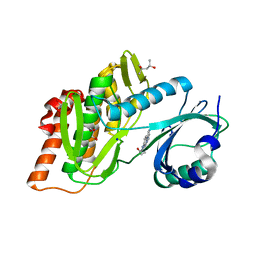 | | Crystal structure of haspin in complex with disubstituted imidazo[1,2- b]pyridazine inhibitor (compound 12) | | Descriptor: | (2~{R})-2-[[3-(2~{H}-indazol-5-yl)imidazo[1,2-b]pyridazin-6-yl]amino]butan-1-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, ... | | Authors: | Chaikuad, A, Bonnet, P, Routier, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of new disubstituted imidazo[1,2- b ]pyridazine derivatives as selective Haspin inhibitors. Synthesis, binding mode and anticancer biological evaluation.
J Enzyme Inhib Med Chem, 35, 2020
|
|
8KFA
 
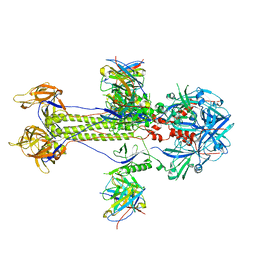 | | Cryo-EM structure of HSV-1 gB with D48 Fab complex | | Descriptor: | D48 heavy chain, D48 light chain, Envelope glycoprotein B | | Authors: | Yang, J, Sun, C, Fang, X, Zeng, M, Liu, Z. | | Deposit date: | 2023-08-15 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | The structure of HSV-1 gB bound to a potent neutralizing antibody reveals a conservative antigenic domain across herpesviruses
hlife, 2023
|
|
6JLS
 
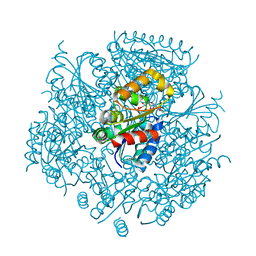 | | Crystal Structure of FMN-dependent Cysteine Decarboxylases TvaF from Thioviridamide Biosynthesis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Putative flavoprotein decarboxylase | | Authors: | Li, J, Lu, J, Wang, H, Zhu, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the FMN-Dependent Cysteine Decarboxylase from Thioviridamide Biosynthesis.
Org.Lett., 21, 2019
|
|
6L30
 
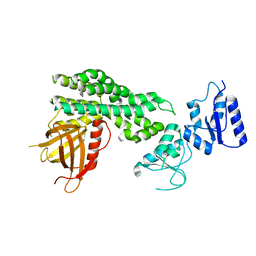 | | Crystal structure of the epithelial cell transforming 2 (ECT2) | | Descriptor: | Protein ECT2 | | Authors: | Chen, Z.C, Chen, M.R, Pan, H, Sun, L.F, Shi, P. | | Deposit date: | 2019-10-07 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and regulation of human epithelial cell transforming 2 protein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7DTE
 
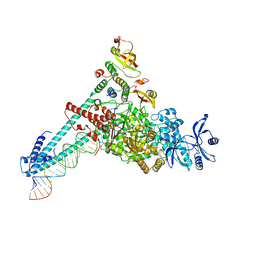 | | SARS-CoV-2 RdRP catalytic complex with T33-1 RNA | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (33-MER), ... | | Authors: | Wang, Q, Gong, P. | | Deposit date: | 2021-01-04 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex.
Cell Rep, 37, 2021
|
|
7VPP
 
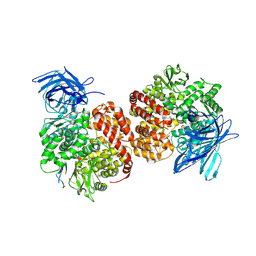 | | Structures of a deltacoronavirus spike protein bound to porcine and human receptors indicate the risk of virus adaptation to humans | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ji, W, Xu, Y, Zhang, S. | | Deposit date: | 2021-10-17 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of a deltacoronavirus spike protein bound to porcine and human receptors.
Nat Commun, 13, 2022
|
|
7VPQ
 
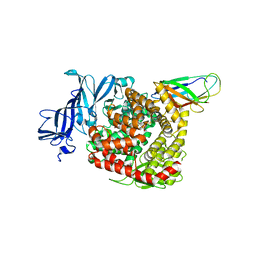 | | Structures of a deltacoronavirus spike protein bound to porcine and human receptors indicate the risk of virus adaptation to humans | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ji, W, Xu, Y, Zhang, S. | | Deposit date: | 2021-10-17 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of a deltacoronavirus spike protein bound to porcine and human receptors.
Nat Commun, 13, 2022
|
|
