7SAD
 
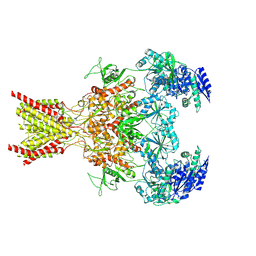 | | Memantine-bound GluN1a-GluN2B NMDA receptors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural insights into binding of therapeutic channel blockers in NMDA receptors.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SAA
 
 | |
7SAC
 
 | | S-(+)-ketamine bound GluN1a-GluN2B NMDA receptors at 3.69 Angstrom resolution | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural insights into binding of therapeutic channel blockers in NMDA receptors.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8BST
 
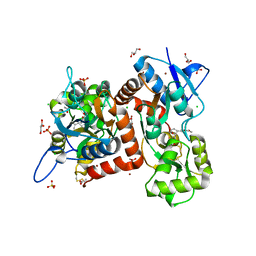 | | Crystal structure of the kainate receptor GluK3-H523A ligand binding domain in complex with kainate at 2.7A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2022-11-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule positive allosteric modulation of homomeric kainate receptors GluK1-3: development of screening assays and insight into GluK3 structure.
Febs J., 291, 2024
|
|
8CKF
 
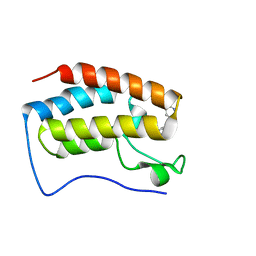 | | Crystal Structure of the first bromodomain of human BRD4 L94C variant in complex with racemic 3,5-dimethylisoxazol ligand | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{S})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Thomas, A.M, McDonough, M.A, Schiedel, M, Conway, S.J. | | Deposit date: | 2023-02-15 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mutate and Conjugate: A Method to Enable Rapid In-Cell Target Validation.
Acs Chem.Biol., 18, 2023
|
|
3C35
 
 | |
3C32
 
 | |
3C31
 
 | |
7P9V
 
 | | Cryo EM structure of System XC- | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
7P9U
 
 | | Cryo EM structure of System XC- in complex with glutamate | | Descriptor: | 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter, GLUTAMIC ACID | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
7OXE
 
 | |
7OYE
 
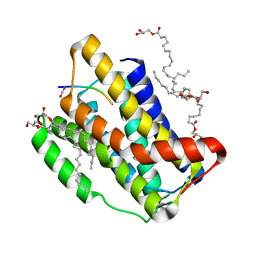 | |
6ZXR
 
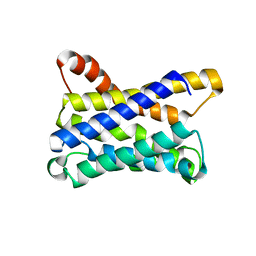 | |
7BC6
 
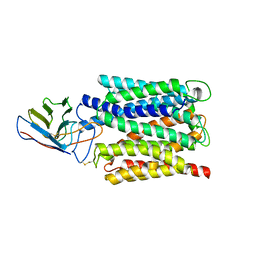 | | Cryo-EM structure of the outward open proton coupled folate transporter at pH 7.5 | | Descriptor: | Proton-coupled folate transporter, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of antifolate recognition and transport by PCFT.
Nature, 595, 2021
|
|
7BC7
 
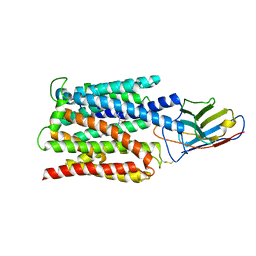 | | Cryo-EM structure of the proton coupled folate transporter at pH 6.0 bound to pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, Proton-coupled folate transporter, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of antifolate recognition and transport by PCFT.
Nature, 595, 2021
|
|
7M6M
 
 | | Full length alpha1 Glycine receptor in presence of 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6N
 
 | |
7M6Q
 
 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 1 | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6R
 
 | |
7M6S
 
 | |
7M6O
 
 | | Full length alpha1 Glycine receptor in presence of 0.1mM Glycine and 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6P
 
 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alphaZ1 | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7NQK
 
 | | Cryo-EM structure of the mammalian peptide transporter PepT2 | | Descriptor: | Solute carrier family 15 member 2, nanobody | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-03-01 | | Release date: | 2021-07-07 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of PepT2 reveals structural basis for proton-coupled peptide and prodrug transport in mammals.
Sci Adv, 7, 2021
|
|
7OG0
 
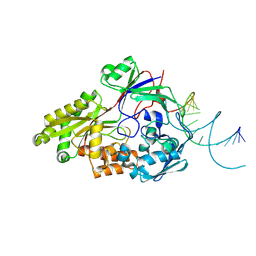 | | Nontypeable Haemophillus influenzae SapA in open and closed conformations, in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
7OFZ
 
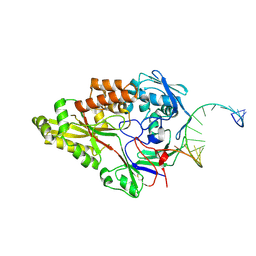 | | Nontypeable Haemophillus influenzae SapA in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
